Annual Report 2018
Division of Epigenomics
Toshikazu Ushijima, Satoshi Yamashita, Hideyuki Takeshima, Naoko Hattori, Masahiro Maeda, Harumi Yamada, Emi Kubo, Naoko Iida, Mika Wakabayashi, Kana Hashimoto, Reiko Nagano, Yuko Miyaji, Naoko Takagi, Asuka Kawachi, Kazuhiro Nishiyama, Takahiro Ebata
Introduction
This Division has been focusing on the epigenetic mechanisms of carcinogenesis, and has identified many aberrantly methylated genes in various cancers, including gastric cancers, esophageal squamous cell carcinomas (ESCCs), neuroblastomas, breast cancers, pancreatic cancers, lung cancers, ovarian cancers, and melanomas. These findings have led to identification of novel tumor-suppressor genes, development of powerful biomarkers, and establishment of the concept of an "epigenetic field for cancerization (epigenetic field defect)". This Division is continuing its activities in 1) revealing induction mechanisms of epigenetic alterations, 2) developing clinically useful biomarkers, and 3) developing an epigenetic cancer therapy.
Research activities
1. Elucidation of Induction Mechanisms of Epigenetic Alterations
Revealing induction mechanisms of epigenetic alterations is very important in understanding cancers. This year, it was revealed that changes of bacterial flora in the colon using antibiotics suppressed tumorigenesis through inhibition of aberrant DNA methylation induced by chronic inflammation. At the same time, it is possible that epigenetic alterations are also accumulated in a cancer microenvironment, and this might enhance cancer development and progression. This year, to address this issue, multiple lines of cancer-associated fibroblasts were established from cancer tissues of gastric cancer patients.
2. Development of Biomarkers
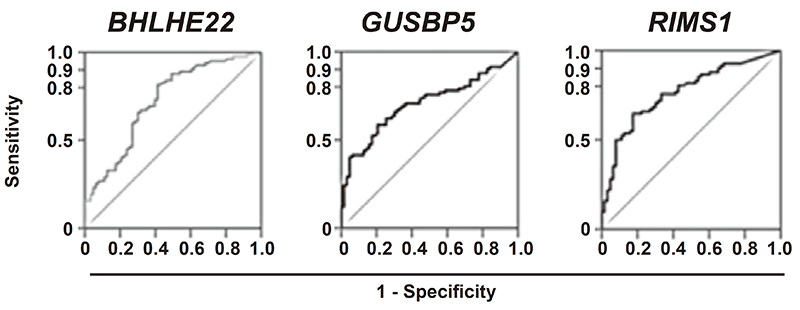
This Division previously has conducted a multicenter prospective cohort study for the prediction of metachronous gastric cancer risk after endoscopic resection, and shown that "the measurement of aberrant DNA methylation accumulated in normal tissues is clinically useful for cancer risk diagnosis". Based on this result, we are now conducting a multicenter prospective cohort study for the prediction of gastric cancer risk in healthy volunteers who underwent eradication of Helicobacter pylori, the almost exclusive cause of gastric cancers. This year, novel risk markers, FLT3, LINC00643, RPRM, JAM2, ELMO1, BHLHE22, RIMS1, GUSBP5, and ZNF93 were developed. Some of these were superior (odds ratios, 5.43-23.41) to a previous marker, miR-124a-3 (Figure 1).
Contamination of normal cells is almost always present in tumor samples and affects their molecular analyses, and development of a biomarker that can estimate the fraction of cancer cells in a tumor DNA sample is important. This year, we identified genomic regions whose DNA methylation levels reflect the fraction of cancer cells in breast cancer and pancreatic cancer samples.
Figure 1. Novel risk markers for gastric cancers

3. Development of epigenetic cancer therapy
DNA demethylation therapy is now expanding from hematological tumors to solid tumors, and stratification of sensitive patients is critically important. This year, a long noncoding RNA, LINC00162, was identified as highly expressed in gastric cancer cell lines sensitive to a DNA demethylating drug. Mechanistically, it was revealed that LINC00162 interacted with an RNA splicing protein, HNRNPH1, and decreased splicing of an anti-apoptotic splicing variant, BCL-XL.
Academia / Training
We have accepted a chief resident from the National Cancer Center Hospital and four student trainees from Tokyo Women's Medical University, Keio University, The University of Tokyo, and Juntendo University. We have also accepted a visiting fellow from the University of Manchester, UK.
Future prospects
Based on these results, this Division will 1) continue multicenter prospective cohort studies for the prediction of gastric cancer risk, 2) conduct the development of epigenetic cancer prevention and therapy, and 3) reveal the molecular mechanisms of how aberrant DNA methylation is induced by chronic inflammation.
List of papers published in 2018
Journal
1. Hattori N, Niwa T, Ishida T, Kobayashi K, Imai T, Mori A, Kimura K, Mori T, Asami Y, Ushijima T. Antibiotics suppress colon tumorigenesis through inhibition of aberrant DNA methylation in an azoxymethane and dextran sulfate sodium colitis model. Cancer Sci, 110:147-156, 2019
2. Abe M, Watanabe K, Shinozaki-Ushiku A, Ushiku T, Abe T, Fujihara Y, Amano Y, Zong L, Wang CP, Kubo E, Inaki R, Kinoshita N, Yamashita S, Takai D, Ushijima T, Nagase T, Hoshi K. Identification of a metastatic lung adenocarcinoma of the palate mucosa through genetic and histopathological analysis: a rare case report and literature review. BMC Cancer, 19:52, 2019
3. Ushijima T, Suzuki H. The Origin of CIMP, At Last. Cancer Cell, 35:165-167, 2019
4. Takeshima H, Ushijima T. Accumulation of genetic and epigenetic alterations in normal cells and cancer risk. NPJ Precis Oncol, 3:7, 2019
5. Sugimoto K, Ito T, Hulbert A, Chen C, Orita H, Maeda M, Moro H, Fukagawa T, Ushijima T, Katai H, Wada R, Sato K, Sakamoto K, Yu W, Considine M, Cope L, Brock MV. DNA methylation genome-wide analysis in remnant and primary gastric cancers. Gastric Cancer, 2019
6. Zong L, Hattori N, Yasukawa Y, Kimura K, Mori A, Seto Y, Ushijima T. LINC00162 confers sensitivity to 5-Aza-2'-deoxycytidine via modulation of an RNA splicing protein, HNRNPH1. Oncogene, 38:5281-5293, 2019
7. Yamaguchi T, Mukai H, Takahashi M, Hara F, Yamauchi C, Yamashita S, Ushijima T. Predictive value of genetic analysis for pathological complete response to preoperative treatment in HER2 positive, HR negative early breast cancer (PASSION trial). Jpn J Clin Oncol, 48:388-391, 2018
8. Tsuchida T, Mano T, Koshi-Mano K, Bannai T, Matsubara T, Yamashita S, Ushijima T, Nagata K, Murayama S, Toda T, Tsuji S, Iwata A. Methylation changes and aberrant expression of FGFR3 in Lewy body disease neurons . Brain Res, 1697:59-66, 2018
9. Yoshida M, Yokota E, Sakuma T, Yamatsuji T, Takigawa N, Ushijima T, Yamamoto T, Fukazawa T, Naomoto Y . Development of an integrated CRISPRi targeting ΔNp63 for treatment of squamous cell carcinoma. Oncotarget, 9:29220-29232, 2018
10. Maeda M, Yamashita S, Shimazu T, Iida N, Takeshima H, Nakajima T, Oda I, Nanjo S, Kusano C, Mori A, Moro H, Yamada H, Tsugane S, Sugiyama T, Sakai Y, Ushijima T. Novel epigenetic markers for gastric cancer risk stratification in individuals after Helicobacter pylori eradication. Gastric Cancer, 21:745-755, 2018
11. Ishihara H, Yamashita S, Fujii S, Tanabe K, Mukai H, Ushijima T. DNA methylation marker to estimate the breast cancer cell fraction in DNA samples. Med Oncol, 35:147, 2018
12. Fukuoka K, Kanemura Y, Shofuda T, Fukushima S, Yamashita S, Narushima D, Kato M, Honda-Kitahara M, Ichikawa H, Kohno T, Sasaki A, Hirato J, Hirose T, Komori T, Satomi K, Yoshida A, Yamasaki K, Nakano Y, Takada A, Nakamura T, Takami H, Matsushita Y, Suzuki T, Nakamura H, Makino K, Sonoda Y, Saito R, Tominaga T, Matsusaka Y, Kobayashi K, Nagane M, Furuta T, Nakada M, Narita Y, Hirose Y, Ohba S, Wada A, Shimizu K, Kurozumi K, Date I, Fukai J, Miyairi Y, Kagawa N, Kawamura A, Yoshida M, Nishida N, Wataya T, Yamaoka M, Tsuyuguchi N, Uda T, Takahashi M, Nakano Y, Akai T, Izumoto S, Nonaka M, Yoshifuji K, Kodama Y, Mano M, Ozawa T, Ramaswamy V, Taylor MD, Ushijima T, Shibui S, Yamasaki M, Arai H, Sakamoto H, Nishikawa R, Ichimura K. Significance of molecular classification of ependymomas: C11orf95-RELA fusion-negative supratentorial ependymomas are a heterogeneous group of tumors. Acta Neuropathol Commun, 6:134, 2018
13. Kunii A, Hara Y, Takenaga M, Hattori N, Fukazawa T, Ushijima T, Yamamoto T, Sakuma T. Three-Component Repurposed Technology for Enhanced Expression: Highly Accumulable Transcriptional Activators via Branched Tag Arrays. CRISPR Journal, 1:337- 347, 2018
14. Ishihara H, Yamashita S, Amano R, Kimura K, Hirakawa K, Ueda T, Murakami Y, Tamori A, Tanabe K, Kawada N, Hagihara A, Ushijima T. Pancreatic Cancer Cell Fraction Estimation in a DNA Sample. Oncology, 95:370-379, 2018
15. Abe M, Zong L, Abe T, Takeshima H, Ji J, Ushijima T, Hoshi K. BRAF inhibitor: a novel therapy for ameloblastoma in mandible. Chin J Cancer Res, 30:677-678, 2018
Book
1. Takeshima H, Yamada H, Ushijima T. Cancer epigenetics: Aberrant DNA methylation in cancer diagnosis and treatment. In: Dammacco F, Silvestris F (eds), Oncogenomics: From Basic Research to Precision Medicine, Academic Press, pp 65-76, 2018
2. Takeshima H, Ushijima T. Mechanisms of DNA methylation changes in cancer. In: Boffetta P, Hainaut P (eds), Encyclopedia of Cancer, 3rd Edition, Academic Press, pp 520-529, 2018
3. Maeda M, Yamada H, Moro H, Ushijima T. Gastric Cancer Risk Prediction Using Epigenetic Alterations Accumulated in Noncancerous Gastric Tissues. In: Shiotani A (ed), Gastric Cancer: With Special Focus on Studies from Japan, Springer, pp 87-98, 2018
