Annual Report 2017
Division of Translational Genomics (Tsukiji Campus)
Takashi Kohno, Hitoshi Ichikawa, Takashi Kubo
Introduction
The Division of Translational Genomics aims to facilitate precision cancer medicine by developing a next-generation sequencing (NGS)-based genomic testing system and advancing its clinical implementation.
Research activities
1. Implementation of "Clinical Sequencing" in Cancer Genome Medicine in Japan
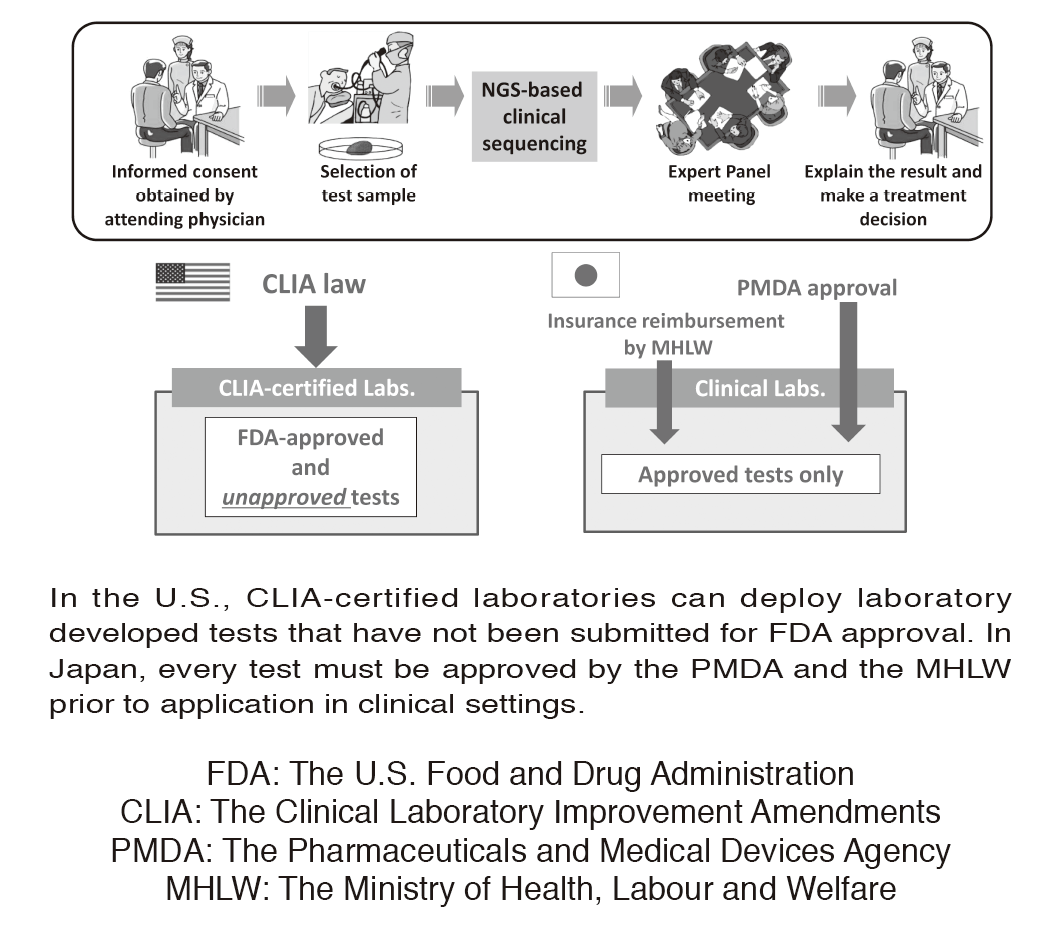
In oncology, actionable mutations (alterations) in cancer-associated genes are critical in terms of a selection of therapeutic approaches. Next-generation sequencing (NGS) of tumor sample DNA (i.e. clinical sequencing) can guide clinical management by providing diagnostic or prognostic data, and facilitating the identification of potential treatment regimens, such as molecular-targeted and immune checkpoint blockade therapies. However, tumor-profiling multiplex gene panel tests have not yet been implemented in routine oncological practice. Due to the lack of CLIA-like regulation, such tests must be approved by the Pharmaceuticals and Medical Devices Agency (PMDA) before insurance reimbursement can be approved by the Japanese Ministry of Health, Labour and Welfare (MHLW) (Figure 1). We are conducting a prospective cohort study to investigate the feasibility and utility of the NGS-based analysis of 114 cancer-associated genes (NCC oncopanel system) in patients with advanced solid tumors. In collaboration with a Japanese diagnostics company, the NCC oncopanel system is now being improved prior to submission for the PMDA approval. This work is being conducted within the context of the SAKIGAKE program of the MHLW.
Figure 1. Differences in laboratory test regulations between the U.S. and Japan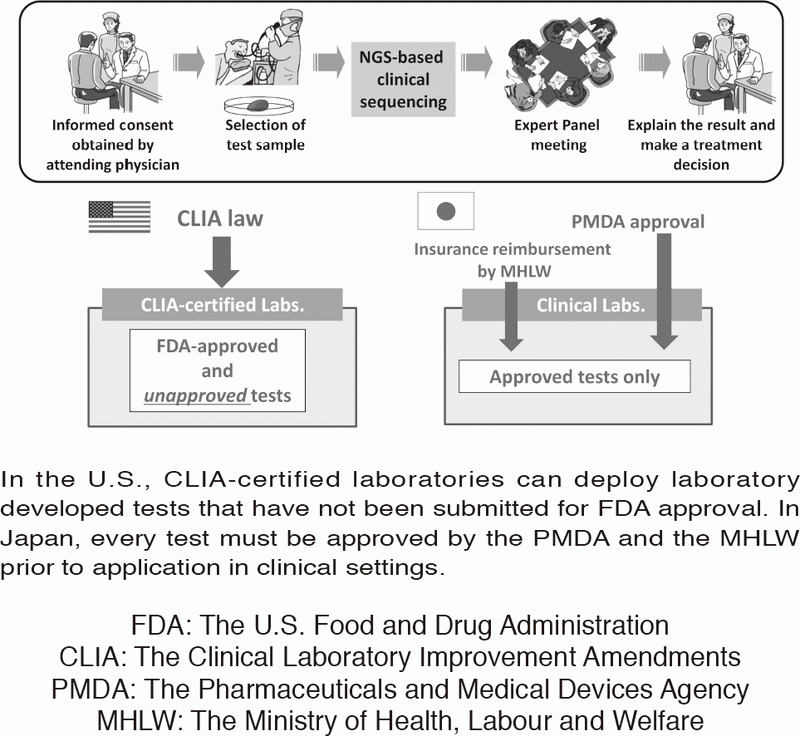
2. NGS-based genomic testing in the NCCH
In cooperation with staff of the Department of Experimental Therapeutics and the Department of Pathology and Clinical Laboratories in the National Cancer Center Hospital (NCCH), prospective clinical studies were performed to examine the feasibility and utility of our NGS-based genomic testing system for patients enrolled into early-phase clinical trials. In the second term of the TOP-GEAR project (May 2016 to March 2018), a laboratory dedicated to NGS analysis compliant with CLIA certification was established in the NCCH, and the NCC Oncopanel system is operated in this laboratory. In the second term, 248 patients suffering from various types of cancer were enrolled and genetic alteration profiles were obtained in 187 patients.
Clinical trials
TOP-GEAR: Trial of Onco-Panel for Gene-profiling to Estimate both Adverse events and Response by cancer treatment (UMIN000011141)
Education
Post-doctoral fellows and chief residents in the NCC were educated through "on-the-job training" in several translational research projects.
Future prospects
In 2018, we plan to validate the clinical utility of the NCC oncopanel system within the Advanced Medical Care B system. The implementation of tumor-profiling multiplex gene panel tests, including NCC oncopanel tests, in cancer genome medicine will be realized in the near future.
In the U.S., CLIA-certified laboratories can deploy laboratory developed tests that have not been submitted for FDA approval. In Japan, every test must be approved by the PMDA and the MHLW prior to application in clinical settings.
FDA: The U.S. Food and Drug Administration
CLIA: The Clinical Laboratory Improvement Amendments
PMDA: The Pharmaceuticals and Medical Devices Agency
MHLW: The Ministry of Health, Labour and Welfare
List of papers published in January 2017 - March 2018
Journal
1. Yoh K, Seto T, Satouchi M, Nishio M, Yamamoto N, Murakami H, Nogami N, Matsumoto S, Kohno T, Tsuta K, Tsuchihara K, Ishii G, Nomura S, Sato A, Ohtsu A, Ohe Y, Goto K. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med, 5:42-50, 2017
2. Hayashi H, Kohno T, Ueno H, Hiraoka N, Kondo S, Saito M, Shimada Y, Ichikawa H, Kato M, Shibata T, Morizane C, Sakamoto Y, Shimada K, Komatsu Y, Sakamoto N, Okusaka T. Utility of Assessing the Number of Mutated KRAS, CDKN2A, TP53, and SMAD4 Genes Using a Targeted Deep Sequencing Assay as a Prognostic Biomarker for Pancreatic Cancer. Pancreas, 46:335-340, 2017
3. Asano N, Yoshida A, Mitani S, Kobayashi E, Shiotani B, Komiyama M, Fujimoto H, Chuman H, Morioka H, Matsumoto M, Nakamura M, Kubo T, Kato M, Kohno T, Kawai A, Kondo T, Ichikawa H. Frequent amplification of receptor tyrosine kinase genes in welldifferentiated/ dedifferentiated liposarcoma. Oncotarget, 8:12941-12952, 2017
4. Ichikawa T, Saruwatari K, Mimaki S, Sugano M, Aokage K, Kojima M, Hishida T, Fujii S, Yoshida J, Kuwata T, Ochiai A, Suzuki K, Tsuboi M, Goto K, Tsuchihara K, Ishii G. Immunohistochemical and genetic characteristics of lung cancer mimicking organizing pneumonia. Lung Cancer, 113:134-139, 2017
5. Suzuki A, Suzuki M, Mizushima-Sugano J, Frith MC, Makalowski W, Kohno T, Sugano S, Tsuchihara K, Suzuki Y. Sequencing and phasing cancer mutations in lung cancers using a long-read portable sequencer. DNA Res, 24:585-596, 2017
6. Yoshida A, Kobayashi E, Kubo T, Kodaira M, Motoi T, Motoi N, Yonemori K, Ohe Y, Watanabe SI, Kawai A, Kohno T, Kishimoto H, Ichikawa H, Hiraoka N. Clinicopathological and molecular characterization of SMARCA4-deficient thoracic sarcomas with comparison to potentially related entities. Mod Pathol, 30:797-809, 2017
7. Nakaoku T, Kohno T, Araki M, Niho S, Chauhan R, Knowles PP, Tsuchihara K, Matsumoto S, Shimada Y, Mimaki S, Ishii G, Ichikawa H, Nagatoishi S, Tsumoto K, Okuno Y, Yoh K, McDonald NQ, Goto K. A secondary RET mutation in the activation loop conferring resistance to vandetanib. Nat Commun, 9:625, 2018
8. Saito M, Saito K, Shiraishi K, Maeda D, Suzuki H, Minamiya Y, Kono K, Kohno T, Goto A. Identification of candidate responders for anti-PD-L1/PD-1 immunotherapy, Rova-T therapy, or EZH2 inhibitory therapy in small-cell lung cancer. Mol Clin Oncol, 8:310-314, 2018
9. Sereewattanawoot S, Suzuki A, Seki M, Sakamoto Y, Kohno T, Sugano S, Tsuchihara K, Suzuki Y. Identification of potential regulatory mutations using multi-omics analysis and haplotyping of lung adenocarcinoma cell lines. Sci Rep, 8:4926, 2018
10. Kohno T. Implementation of "clinical sequencing" in cancer genome medicine in Japan. Cancer Sci, 109:507-512, 2018
