Annual Report 2017
Division of Epigenomics
Toshikazu Ushijima, Satoshi Yamashita, Hideyuki Takeshima, Naoko Hattori, Masahiro Maeda, Hiroshi Moro, Naoko Iida, Emi Kubo, Harumi Yamada, Hironori Takamatsu, Hiroki Ishihara, Asuka Kawachi, Zong Liang, Mika Wakabayashi, Akiko Mori, Kana Kimura, Reiko Nagano, Yuko Miyaji, Naoko Kobayashi, Aya Nakajima, Masanobu Abe
Introduction
The Division of Epigenomics has been focusing on the epigenetic mechanisms of carcinogenesis, and has identified many aberrantly methylated genes in various cancers, including gastric cancers, esophageal squamous cell carcinomas (ESCCs), neuroblastomas, breast cancers, pancreatic cancers, lung cancers, ovarian cancers, and melanomas. These findings have led to identification of novel tumor-suppressor genes, the development of powerful biomarkers, and the establishment of the concept of an "epigenetic field for cancerization (epigenetic field defect)". This division continues its activity in 1) developing clinically useful biomarkers, a novel approach to cancer prevention, and epigenetic therapy and 2) revealing molecular mechanisms of induction of aberrant DNA methylation.
Research activities
1. Identification of Novel Epigenetic Alterations
Identification of tumor-suppressor genes inactivated by epigenetic alterations, such as aberrant DNA methylation, is important. This year, a web tool, which can be used for the analysis of DNA methylation data obtained by BeadArray technologies, was developed.
2. Development of Biomarkers
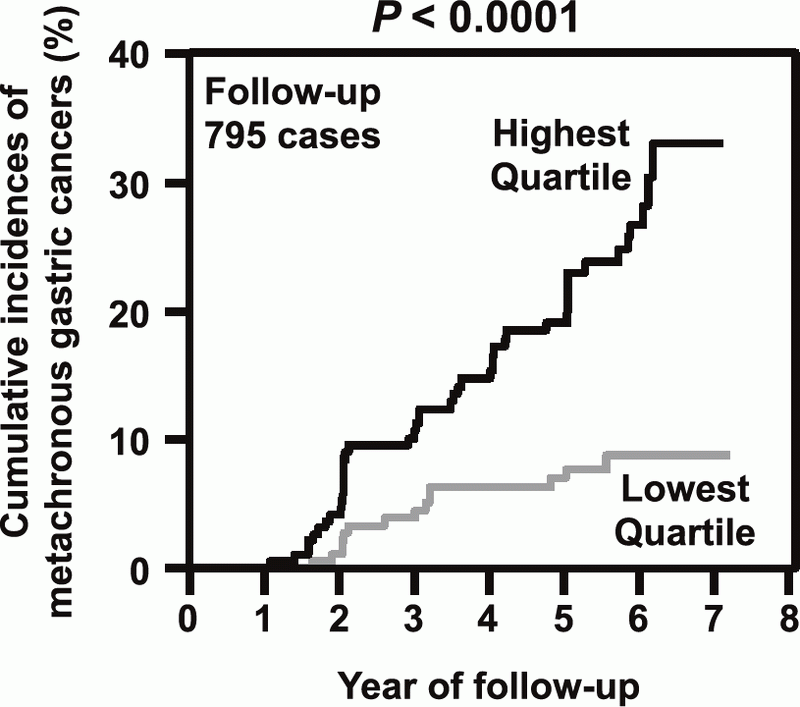
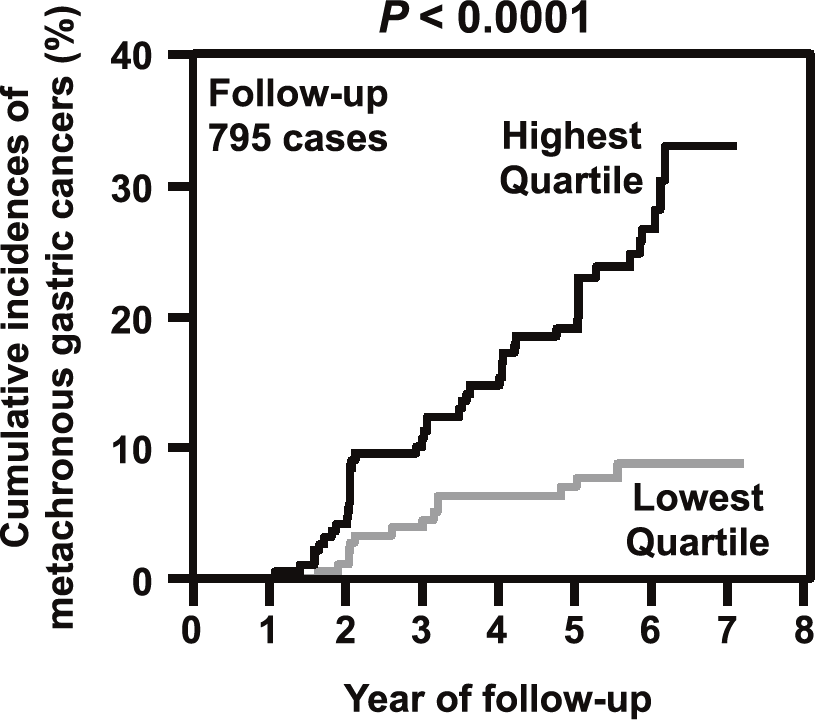
This division previously revealed that the degree of accumulated aberrant DNA methylation in normal-appearing gastric mucosae is expected to be a useful diagnostic marker to predict gastric cancer risk. To bring this concept into clinical practice, a multicenter prospective cohort study has been conducted for the prediction of metachronous gastric cancer risk after
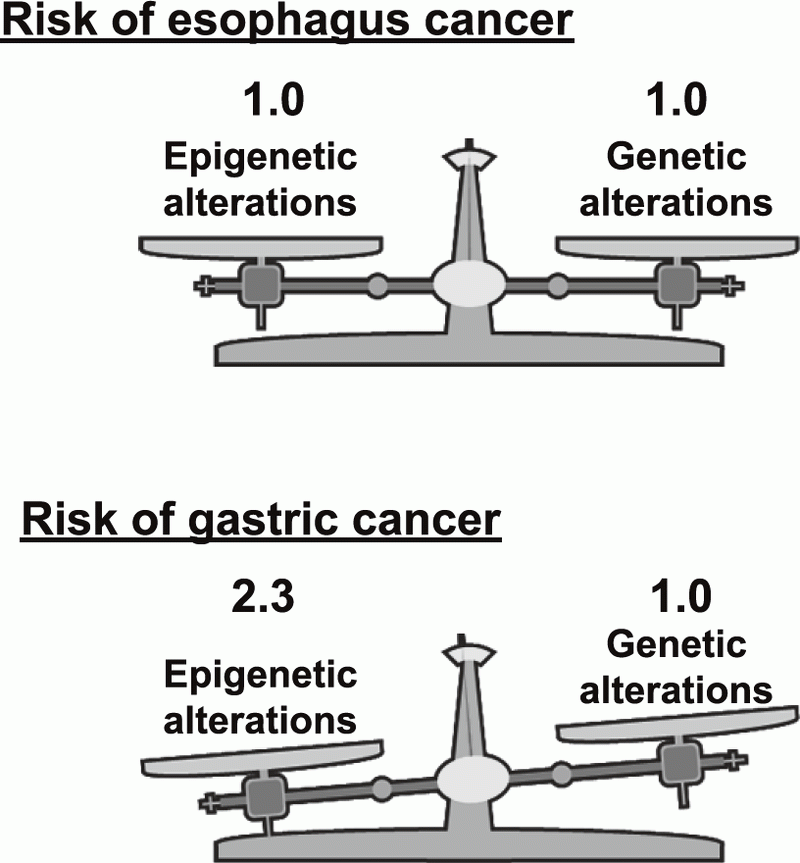
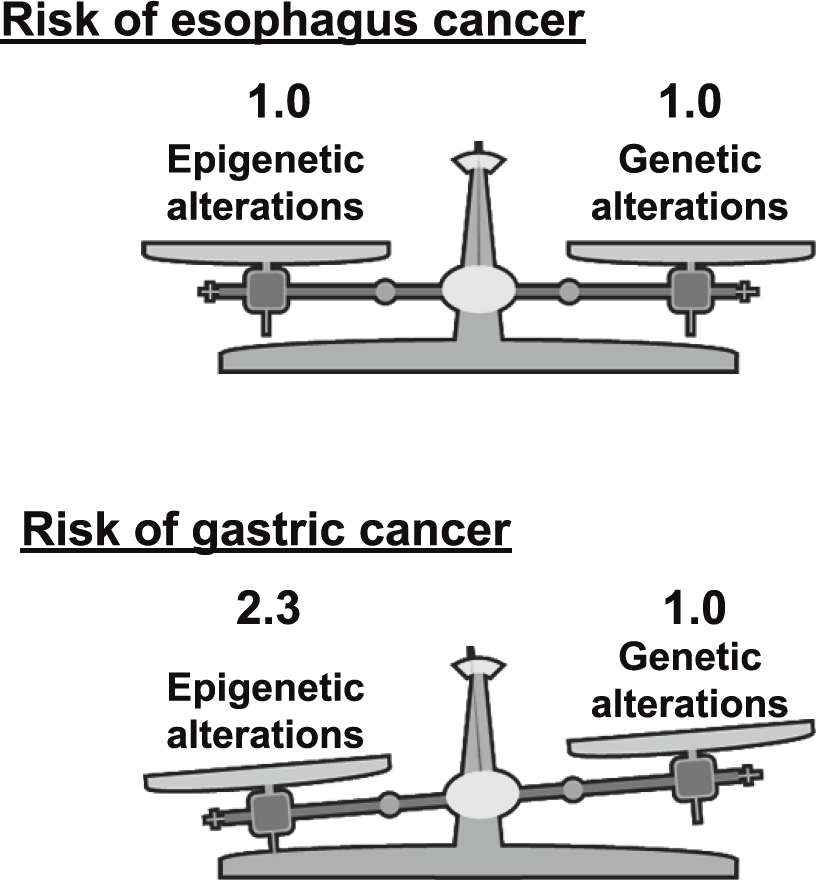
endoscopic resection. This year, a clear result of the prospective study, "the measurement of aberrant DNA methylation accumulated in normal tissues is clinically useful for cancer risk diagnosis", was published (Figure 1). In addition, a novel technology that can detect mutations of frequencies at the 10-6/bp level accumulated in normal tissues was developed. Using this technology, it was shown that, in the stomach, epigenetic alterations have a higher impact on cancer risk than genetic alterations (Figure 2).
On the basis of these results, we are conducting a multicenter prospective cohort study for the prediction of gastric cancer risk in healthy volunteers who underwent eradication of Helicobacter pylori, the almost exclusive cause of gastric cancers. This year, an initial methylation profile of healthy volunteers was determined.
It was also shown that DNA methylation status of HSD17B4 is useful to predict therapeutic efficacy of HER2-targeting therapy in HER2-
positive breast cancer patients.
3. Development of epigenetic cancer therapy
As a strategy against cancers resistant to anticancer drugs or molecular target drugs, a combination therapy involving an epigenetic drug can be useful. This year, the usefulness of a combination of a BET inhibitor and a mutant BRAF inhibitor, vemurafenib, for the treatment of colon cancers with BRAF mutations was shown.
Future prospects
On the basis of the results mentioned above, this division will 1) continue multicenter prospective cohort studies for the prediction of gastric cancer risk, 2) conduct the development of epigenetic cancer prevention and therapy, and 3) reveal the molecular mechanisms of how aberrant DNA methylation is induced by chronic inflammation.
List of papers published in January 2017 - March 2018
Journal
1. Maeda M, Moro H, Ushijima T. Mechanisms for the induction of gastric cancer by Helicobacter pylori infection: aberrant DNA methylation pathway. Gastric Cancer, 20:8-15, 2017
2. Nakamura T, Yamashita S, Fukumura K, Nakabayashi J, Tanaka K, Tamura K, Tateishi K, Kinoshita M, Fukushima S, Takami H, Fukuoka K, Yamazaki K, Matsushita Y, Ohno M, Miyakita Y, Shibui S, Kubo A, Shuto T, Kocialkowski S, Yamanaka S, Mukasa A, Sasayama T, Mishima K, Maehara T, Kawahara N, Nagane M, Narita Y, Mano H, Ushijima T, Ichimura K. Genome-wide DNA methylation profiling identifies primary central nervous system lymphoma as a distinct entity different from systemic diffuse large B-cell lymphoma. Acta Neuropathol, 133:321-324, 2017
3. Fukushima S, Yamashita S, Kobayashi H, Takami H, Fukuoka K, Nakamura T, Yamasaki K, Matsushita Y, Nakamura H, Totoki Y, Kato M, Suzuki T, Mishima K, Yanagisawa T, Mukasa A, Saito N, Kanamori M, Kumabe T, Tominaga T, Nagane M, Iuchi T, Yoshimoto K, Mizoguchi M, Tamura K, Sakai K, Sugiyama K, Nakada M, Yokogami K, Takeshima H, Kanemura Y, Matsuda M, Matsumura A, Kurozumi K, Ueki K, Nonaka M, Asai A, Kawahara N, Hirose Y, Takayama T, Nakazato Y, Narita Y, Shibata T, Matsutani M, Ushijima T, Nishikawa R, Ichimura K. Genome-wide methylation profiles in primary intracranial germ cell tumors indicate a primordial germ cell origin for germinomas. Acta Neuropathol, 133:445-462, 2017
4. Yoshida S, Yamashita S, Niwa T, Mori A, Ito S, Ichinose M, Ushijima T. Epigenetic inactivation of FAT4 contributes to gastric field cancerization. Gastric Cancer, 20:136-145, 2017
5. Narita M, Shimura E, Nagasawa A, Aiuchi T, Suda Y, Hamada Y, Ikegami D, Iwasawa C, Arakawa K, Igarashi K, Kuzumaki N, Yoshioka Y, Ochiya T, Takeshima H, Ushijima T, Narita M. Chronic treatment of non-small-cell lung cancer cells with gefitinib leads to an epigenetic loss of epithelial properties associated with reductions in microRNA-155 and -200c. PLoS One, 12:e0172115, 2017
6. Fujii S, Yamashita S, Yamaguchi T, Takahashi M, Hozumi Y, Ushijima T, Mukai H. Pathological complete response of HER2-positive breast cancer to trastuzumab and chemotherapy can be predicted by HSD17B4 methylation. Oncotarget, 8:19039-19048, 2017
7. Ushijima T. Precision epigenetic risk diagnosis after mass eradication of H. pylori. Rinsho Hyoka, 45:128-129, 2017
8. Nakamura Y, Hattori N, Iida N, Yamashita S, Mori A, Kimura K, Yoshino T, Ushijima T. Targeting of super-enhancers and mutant BRAF can suppress growth of BRAF-mutant colon cancer cells via repression of MAPK signaling pathway. Cancer Lett, 402:100-109, 2017
9. Yamashita S, Iida N, Takeshima H, Hattori N, Maeda M, Kishino T, Nagano R, Shimazu T, Tsugane S, Ushijima T. A novel method to quantify base substitution mutations at the 10-6 per bp level in DNA samples. Cancer Lett, 403:152-158, 2017
10. Maeda M, Nakajima T, Oda I, Shimazu T, Yamamichi N, Maekita T, Asada K, Yokoi C, Ando T, Yoshida T, Nanjo S, Fujishiro M, Gotoda T, Ichinose M, Ushijima T. High impact of methylation accumulation on metachronous gastric cancer: 5-year follow-up of a multicentre prospective cohort study. Gut, 66:1721-1723, 2017
11. Padmanabhan N, Ushijima T, Tan P. How to stomach an epigenetic insult: the gastric cancer epigenome. Nat Rev Gastroenterol Hepatol, 14:467-478, 2017
12. West AC, Tang K, Tye H, Yu L, Deng N, Najdovska M, Lin SJ, Balic JJ, Okochi-Takada E, McGuirk P, Keogh B, McCormack W, Bhathal PS, Reilly M, Oshima M, Ushijima T, Tan P, Jenkins BJ. Identification of a TLR2-regulated gene signature associated with tumor cell growth in gastric cancer. Oncogene, 36:5134-5144, 2017
13. Tsubota S, Kishida S, Shimamura T, Ohira M, Yamashita S, Cao D, Kiyonari S, Ushijima T, Kadomatsu K. PRC2-Mediated Transcriptomic Alterations at the Embryonic Stage Govern Tumorigenesis and Clinical Outcome in MYCN-Driven Neuroblastoma. Cancer Res, 77:5259-5271, 2017
14. Mano T, Nagata K, Nonaka T, Tarutani A, Imamura T, Hashimoto T, Bannai T, Koshi-Mano K, Tsuchida T, Ohtomo R, Takahashi-Fujigasaki J, Yamashita S, Ohyagi Y, Yamasaki R, Tsuji S, Tamaoka A, Ikeuchi T, Saido TC, Iwatsubo T, Ushijima T, Murayama S, Hasegawa M, Iwata A. Neuron-specific methylome analysis reveals epigenetic regulation and tau-related dysfunction of BRCA1 in Alzheimer's disease. Proc Natl Acad Sci U S A, 114:E9645-E9654, 2017
15. Hironaka-Mitsuhashi A, Matsuzaki J, Takahashi RU, Yoshida M, Nezu Y, Yamamoto Y, Shiino S, Kinoshita T, Ushijima T, Hiraoka N, Shimizu C, Tamura K, Ochiya T. A tissue microRNA signature that predicts the prognosis of breast cancer in young women. PLoS One, 12:e0187638, 2017
16. Hashimoto T, Yamashita S, Yoshida H, Taniguchi H, Ushijima T, Yamada T, Saito Y, Ochiai A, Sekine S, Hiraoka N. WNT Pathway Gene Mutations Are Associated With the Presence of Dysplasia in Colorectal Sessile Serrated Adenoma/Polyps. Am J Surg Pathol, 41:1188-1197, 2017
17. Gao D, Herman JG, Cui H, Jen J, Fuks F, Brock MV, Ushijima T, Croce C, Akiyama Y, Guo M. Meeting Report of the Fifth International Cancer Epigenetics Conference in Beijing, China, October 2016. Epigenomics, 9:937-941, 2017
18. Takeshima H, Niwa T, Toyoda T, Wakabayashi M, Yamashita S, Ushijima T. Degree of methylation burden is determined by the exposure period to carcinogenic factors. Cancer Sci, 108:316-321, 2017
19. Muhammad JS, Nanjo S, Ando T, Yamashita S, Maekita T, Ushijima T, Tabuchi Y, Sugiyama T. Autophagy impairment by Helicobacter pylori-induced methylation silencing of MAP1LC3Av1 promotes gastric carcinogenesis. Int J Cancer, 140:2272-2283, 2017
20. Yamashita S, Kishino T, Takahashi T, Shimazu T, Charvat H, Kakugawa Y, Nakajima T, Lee YC, Iida N, Maeda M, Hattori N, Takeshima H, Nagano R, Oda I, Tsugane S, Wu MS, Ushijima T. Genetic and epigenetic alterations in normal tissues have differential impacts on cancer risk among tissues. Proc Natl Acad Sci U S A, 115:1328-1333, 2018
21. Hattori N, Ushijima T. Analysis of DNA Methylation in Tissues Exposed to Inflammation. Methods Mol Biol, 1725:185-199, 2018
22. Iida N, Okuda Y, Ogasawara O, Yamashita S, Takeshima H, Ushijima T. MACON: a web tool for computing DNA methylation data obtained by the Illumina Infinium Human DNA methylation BeadArray. Epigenomics, 10:249-258, 2018
23. Okochi-Takada E, Hattori N, Ito A, Niwa T, Wakabayashi M, Kimura K, Yoshida M, Ushijima T. Establishment of a high-throughput detection system for DNA demethylating agents. Epigenetics, 13:147-155, 2018
24. Herceg Z, Ghantous A, Wild CP, Sklias A, Casati L, Duthie SJ, Fry R, Issa JP, Kellermayer R, Koturbash I, Kondo Y, Lepeule J, Lima SCS, Marsit CJ, Rakyan V, Saffery R, Taylor JA, Teschendorff AE, Ushijima T, Vineis P, Walker CL, Waterland RA, Wiemels J, Ambatipudi S, Degli Esposti D, Hernandez-Vargas H. Roadmap for investigating epigenome deregulation and environmental origins of cancer. Int J Cancer, 142:2018
Book
1. Hattori N, Ushijima T. Analysis of gene-specific DNA methylation. In: Tollefsbol T (ed), Handbook of Epigenetics 2nd Edition: The New Molecular and Medical Genetics, Academic Press, pp 113-123, 2017
2. Takeshima H, Ushijima T. Future perspective of DNA and histone methylation as cancer targets. In: Kaneda A, Tsukada Y (eds), DNA and Histone Methylation as Cancer Targets, Humana Press, pp 607-622, 2017
