Annual Report 2017
Division of Molecular Modification and Cancer Biology
Ryuji Hamamoto, Syuzo Kaneko, Masaaki Komatsu, Masayoshi Yamada, Masamichi Takahashi, Kazuma Kobayashi, Kyoko Fujioka, Noriko Ikawa, Hiroko Kondo, Shigemi Yamada, Michihiro Sakai, Kruthi Suvarna, Hidenori Machino, Sangchul Kim, Yuichiro Yoshioka
Introduction
In recent years, it is reported that abnormality of molecular modification including protein modification and the nucleic acid modification plays a critical role in human tumorigenesis, and the enzyme group controlling abnormal molecular modification is considered as an important target for cancer treatment. Since we clarified the molecular mechanism of how abnormal histone methylation causes human tumorigenesis ahead of the world (Hamamoto et al., Nature Cell Biology, 6, 731-740 [2004]), we have continuously been studying the significance of abnormal histone methylation in human cancer. Consequently, we have already identified a number of protein methyltransferases and demethylases as molecular targets for cancer therapy (Hamamoto et al., Nature Reviews Cancer, 15, 110-124 [2015]). In the Division of Molecular Modification and Cancer Biology, we try to elucidate the molecular mechanism of how abnormal modification causes human tumorigenesis, in particular focusing on protein methylation, and apply our findings for development of novel anti-cancer drugs. In addition, we introduced artificial intelligence (AI) technologies into a medical study during early stages of the development of medical AI in Japan, and develop the integrated cancer medical system using the AI technology intended to apply to Precision Medicine. We now organize one of the research projects of CREST, a national project, called "Development of integrated Medical System for Diagnosis and Treatment of Cancer by Artificial Intelligence".
Research activities
1. Development of the next generation ChIP-seq method that intends application to Precision Medicine
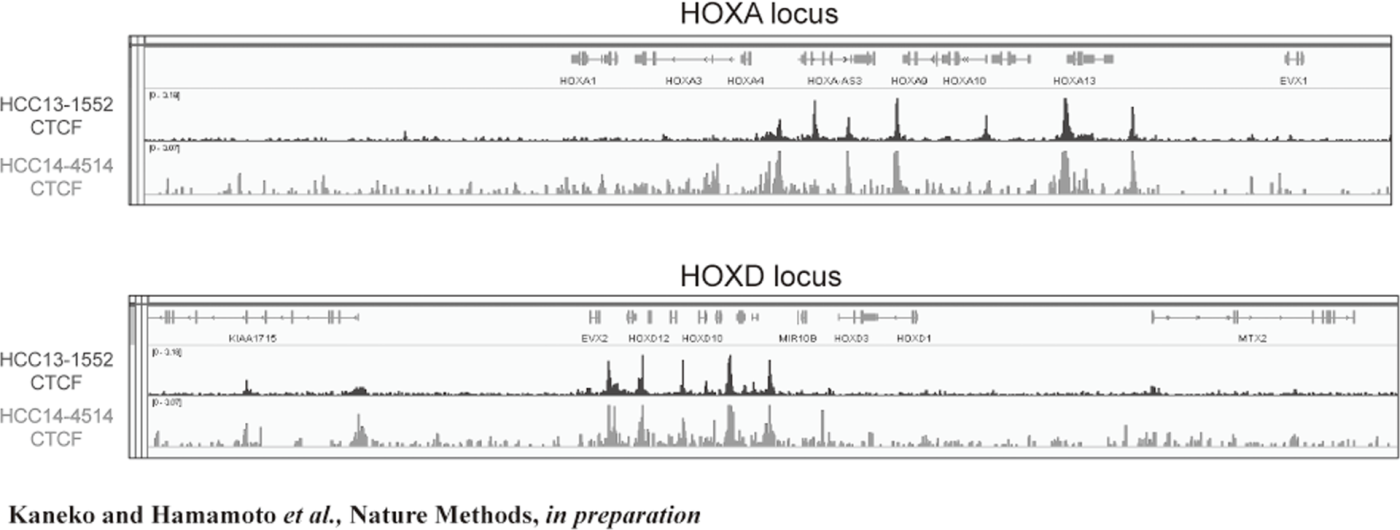
In order to establish the platform to conduct multi-omics analysis using machine learning and deep learning, we tried to develop the next-generation ChIP-seq method. Consequently, we succeeded in developing a system, which enables the ChIP-seq analysis of formalin-fixed, paraffin-embedded (FFPE) samples effectively. On the basis of this system, we successfully identified not only status of histone modifications (H3K27 acety-lation, H3K4 tri-methylation) (Figure 1) but also the identification of the binding sites of transcription factor CCCTC-binding Factor (CTCF). This is the first case, which succeeds for the identification of the transcription factor binding sites by ChIP-seq analysis using the FFPE sample (Figure 2). In this system, we introduced the human general-purpose robot LabDroid, which enabled acquisition of highly robust data.
2. Development of highly precise image diagnosis devices and image diagnosis methods by AI technology
We continuously worked on the development of a real-time endoscope diagnosis support system to detect colon cancer and precancerous lesions using Deep Learning. As a result, regarding an upheaval type lesion, we succeeded in the development of the system to examine 98% of sensitivity, specific degree 99% and plus, and to boast of precision of rate 98.8%. These results indicate that our own developed system shows the world's top level quality. We are now preparing for a submission of the article, and consulting with the Pharmaceuticals and Medical Devices Agency (PMDA) to use it in a clinical situation.
As for the radiation image analysis, we were able to analyze chronological change of the image characteristic quantity, which we could not catch by the change of a simple tumor size, by automated Radiomics analysis system cyclopaedically and quantitatively. Furthermore, we succeeded in the construction of a detector which improved precision selectively for a brain tumor region by doing fine tuning in the last output layer of 3D-Unet using the segmentation.
Education
Five graduate school students from the University of Tokyo, Department of Medicine, and one graduate school student from Kyorin University, were trained in our division. One of our staff members won the National Cancer Center Research Institute (NCCRI) director award.
Future prospects
1) To develop the analytical method of multi-omics data using the AI technology to apply to Precision Medicine
2) To analyze the chromatin structure of clinical tissues using ATAC-seq and ChIP-seq methods, and examine the possibility of clinical application
3) To develop an integrated medical system for cancer treatment using the AI technology
Figure 1. FFPE ChIP-Seq (H3K4me3)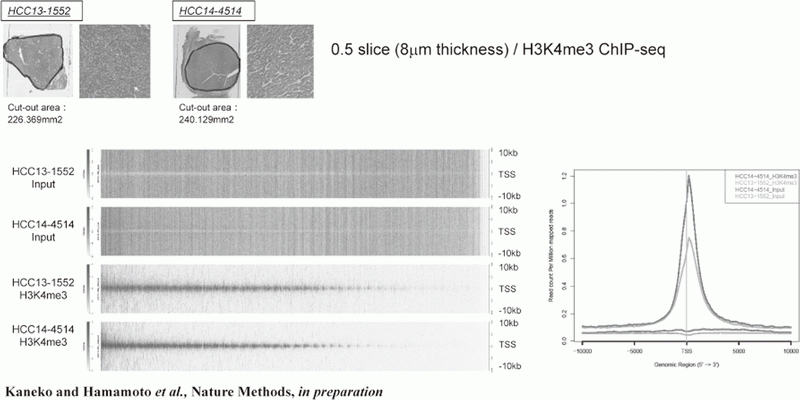
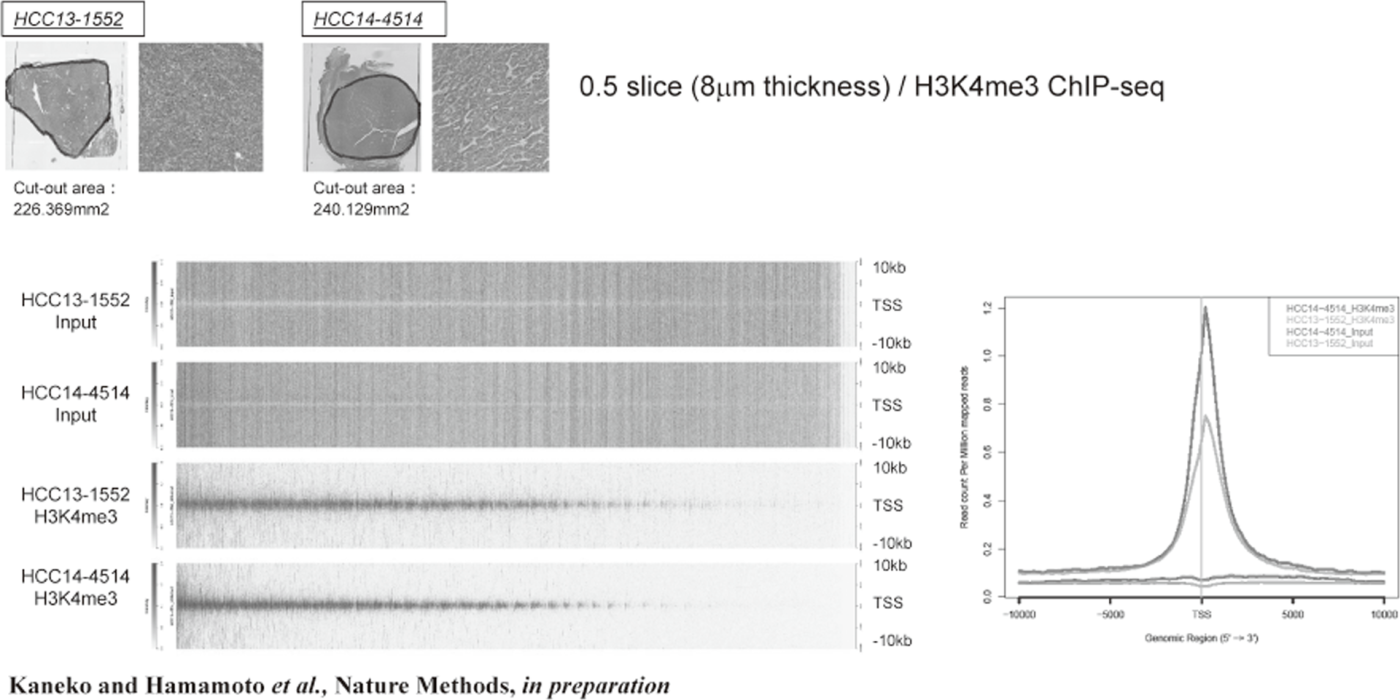
Figure 2. FFPE ChIP-Seq (CTCF)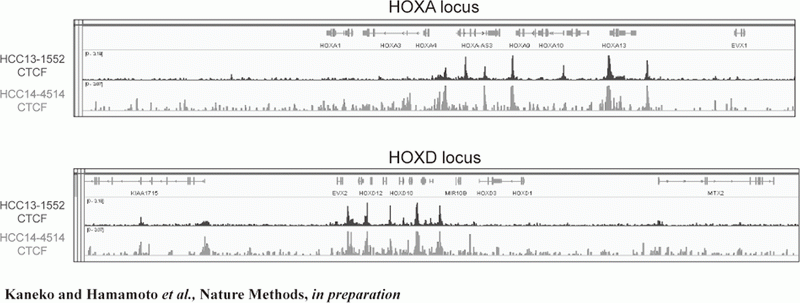
List of papers published in January 2017 - March 2018
Journal
1. Yoshioka Y, Suzuki T, Matsuo Y, Tsurita G, Watanabe T, Dohmae N, Nakamura Y, Hamamoto R. Protein lysine methyltransferase SMYD3 is involved in tumorigenesis through regulation of HER2 homodimerization. Cancer Med, 6:1665-1672, 2017
2. Chiyoda T, Hart PC, Eckert MA, McGregor SM, Lastra RR, Hamamoto R, Nakamura Y, Yamada SD, Olopade OI, Lengyel E, Romero IL. Loss of BRCA1 in the Cells of Origin of Ovarian Cancer Induces Glycolysis: A Window of Opportunity for Ovarian Cancer Chemoprevention. Cancer Prev Res (Phila), 10:255-266, 2017
3. Toyokawa G, Takada K, Okamoto T, Kozuma Y, Matsubara T, Haratake N, Akamine T, Takamori S, Katsura M, Shoji F, Hamamoto R, Oda Y, Maehara Y. Elevated Metabolic Activity on (18)F-FDG PET/CT Is Associated with the Expression of EZH2 in Non-small Cell Lung Cancer. Anticancer Res, 37:1393-1401, 2017
4. Wang R, Deng X, Yoshioka Y, Vougiouklakis T, Park JH, Suzuki T, Dohmae N, Ueda K, Hamamoto R, Nakamura Y. Effects of SMYD2-mediated EML4-ALK methylation on the signaling pathway and growth in non-small-cell lung cancer cells. Cancer Sci, 108:1203-1209, 2017
5. Kunizaki M, Fukuda A, Wakata K, Tominaga T, Nonaka T, Miyazaki T, Matsumoto K, Sumida Y, Hidaka S, Yasutake T, Sawai T, Hamamoto R, Nanashima A, Nagayasu T. Clinical Significance of Serum p53 Antibody in the Early Detection and Poor Prognosis of Gastric Cancer. Anticancer Res, 37:1979-1984, 2017
6. Oki S, Sone K, Oda K, Hamamoto R, Ikemura M, Maeda D, Takeuchi M, Tanikawa M, Mori-Uchino M, Nagasaka K, Miyasaka A, Kashiyama T, Ikeda Y, Arimoto T, Kuramoto H, Wada-Hiraike O, Kawana K, Fukayama M, Osuga Y, Fujii T. Oncogenic histone methyltransferase EZH2: A novel prognostic marker with therapeutic potential in endometrial cancer. Oncotarget, 8:40402-40411, 2017
7. Saloura V, Vougiouklakis T, Zewde M, Deng X, Kiyotani K, Park JH, Matsuo Y, Lingen M, Suzuki T, Dohmae N, Hamamoto R, Nakamura Y. WHSC1L1-mediated EGFR mono-methylation enhances the cytoplasmic and nuclear oncogenic activity of EGFR in head and neck cancer. Sci Rep, 7:40664, 2017
8. Deng X, Hamamoto R, Vougiouklakis T, Wang R, Yoshioka Y, Suzuki T, Dohmae N, Matsuo Y, Park JH, Nakamura Y. Critical roles of SMYD2-mediated beta-catenin methylation for nuclear translocation and activation of Wnt signaling. Oncotarget, 8:55837-55847, 2017
9. Lee CH, Yu JR, Kumar S, Jin Y, Kaneko S, Hamilton AD, Reinberg D. Allosteric activation dictates PRC2 activity independent of its recruitment to chromatin. bioRxiv, 2017
10. Kunizaki M, Tominaga T, Wakata K, Miyazaki T, Matsumoto K, Sumida Y, Hidaka S, Yamasaki T, Yasutake T, Sawai T, Hamamoto R, Nanashima A, Nagayasu T. Clinical significance of the C-reactive protein-to-albumin ratio for the prognosis of patients with esophageal squamous cell carcinoma. Mol Clin Oncol, 8:370-374, 2018
11. Kunizaki M, Hamasaki K, Wakata K, Tobinaga S, Sumida Y, Hidaka S, Yasutake T, Miyazaki T, Matsumoto K, Yamasaki T, Sawai T, Hamamoto R, Nanashima A, Nagayasu T. Clinical Value of Serum p53 Antibody in the Diagnosis and Prognosis of Esophageal Squamous Cell Carcinoma. Anticancer Res, 38:1807-1813, 2018
12. Piao L, Fujioka K, Nakakido M, Hamamoto R. Regulation of poly(ADP-Ribose) polymerase 1 functions by post-translational modifications. Front Biosci (Landmark Ed), 23:13-26, 2018
