Annual Report 2018
Division of Biomarker Discovery (Kashiwa Campus)
Atsushi Ochiai, Syuichi Mitsunaga, Tomofumi Miura, Kazuyoshi Yanagihara, Yuki Iino, Akiko Nagatsuma, Aoi Sukeda, Chisako Yamauchi, Yuka Nakamura, Aya Endo, Misato ohasi, Mariko Yajima, Michiko Hamamoto, Teruo Komatsu, Takahiro Iimura, Sachiko Fukuda, Motoko Suzaki
Introduction
In the translational research field for the exploration of biomarkers, we have been exploring biomarkers which enable the prediction of the prognoses of cancer patients and biological figures of cancer, and constructing companion diagnoses that relate to the clinical studies with the aim to achieve personalized (optimized) medicine.
The Team and What We Do
In the translational research field for the exploration of biomarkers, we have been exploring biomarkers which enable the prediction of the prognoses of cancer patients and biological figures of cancer, and constructing companion diagnoses that relate to the clinical studies with the aim to achieve personalized (optimized) medicine.
Research activities
The research achievements of this year are summarized into the following four points:
1) In the research and development of molecular targeted drugs, it is very important to develop a target that is characteristic to the particular cancer. It has been known that cancerous cachexia takes place with a high frequency as the clinicopathological feature of pancreatic cancer. The metabolic mechanisms in the patients with pancreatic cancer change greatly including the increases in the lactic acid level and ketone body level in their blood. We studied the expression of monocarboxylate receptors which enable the cancer or stromal cells to utilize the lactic acid and ketone body and their clinicopathological relation using 240 cases of consecutively resected surgical specimens. As a result, it was found that the high expression of monocarboxylate 1 (MCT1) in cancer cells relates to the good prognosis of the patient, and high expression of MCT4 in the stromal fibroblast relates to the poor prognosis. These results showed that cancerous tissues consisting of cancer cells and fibroblast cells have molecular activities that correspond to the dynamic change in their metabolism change; therefore, those molecular activities may be future novel biomarkers or therapeutic targets.
2) We transplanted the cultured human pancreatic cancer cells to the sciatic nerve of immunodeficient mice with the aim to create a model which can reproduce the cancer pain of pancreatic cancer. In the study, we found that the inflammation in dorsal root ganglia (DRG) changes the pain threshold resulting in the strong pain felt by cancer patients (allodynia). We confirmed that this pain in the animal model is actually taking place in human patients by comparing the degrees of abdominal perception (stimulation with 2,000 Hz and utilization of a pressure sensor) and nerve infiltration in 43 pancreatic cancer patients who underwent operations after examination. As a result, it was shown that in cases in which the nerve infiltration was detected by CT, the pain threshold thereof was changing; thus, the likelihood of the animal model, in which nerve inflammation is caused by the nerve infiltration of the pancreatic cancer, strongly correlating to the cancer pain. Additionally, we investigated that the activation state of ghrelin in the blood of patients with pancreatic cancer was compared to their anorexia, and the condition of the activated ghrelin was strongly correlated with the appetite of the patients.
3) We have supported the studies in the biobank, establishments of cancer xenografts (patientderived xenograft: PDX) and novel cell line studies on the establishment of xenografts as well as novel cell lines (the DEF Study) that have been performed in the National Cancer Center Hospital East. In this study, a total of 250 cases were used, 233 cases of surgical specimens were xenografted resulting in 114 cases of successful engraftment. Among them, 40 cases were diagnosed as a cancer by histopathological examination and 35 strains were established as PDX. As cell lines, 7 cell lines were obtained out of 17 ascitic fluid specimens; 25 cell lines were obtained out of surgical specimens and xenograft; thus, a total of 32 strains of cultured cell were established. Additionally, as a project related to the biobank in the National Cancer Center, we collected serums from patients with multiple cancers caused by miRNAs to cooperate with the research.
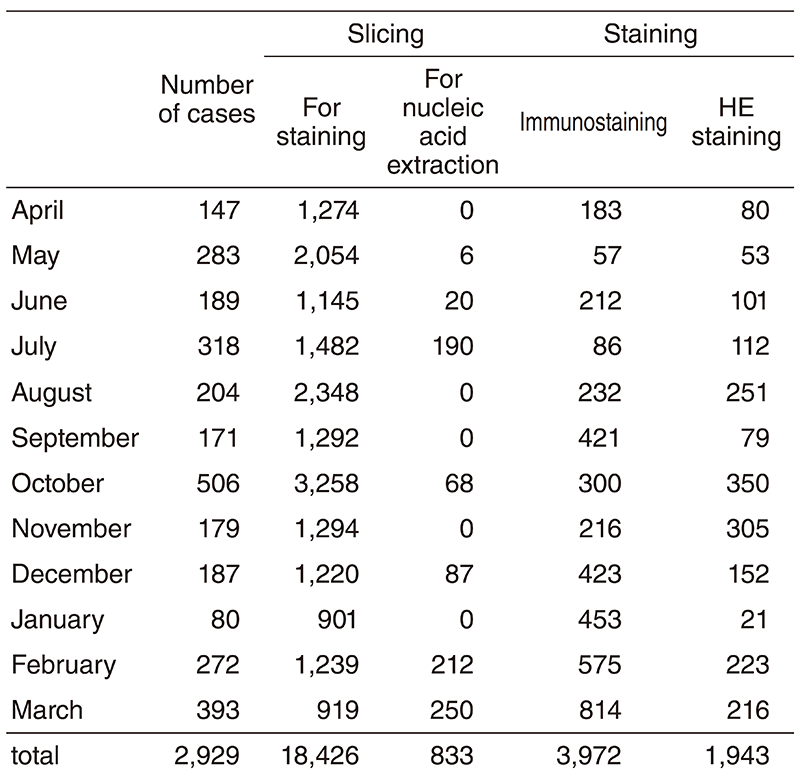
4) Regarding the preparation of PDX from tissues of pathological specimens, which has been conducted in the Exploratory Oncology Research and Clinical Trial Center, we prepared pathological slices from surgical and animal model specimens and instructed researchers about the immunostaining procedure and how to evaluate the results of immunostaining. For the purpose of supporting the study using pathological specimens, we supported the preparation of specimens as shown in the table below.
Table 1. Slicing and dyeing
(cumulative totals April 2018 - March 2019)

Future prospects
Since genome-based personalized (optimal) medicine has been started from this year, further biomarkers including the target in cancer and the prediction of the effectiveness of treatment and patients' survival are required. The Division of Biomarker Discovery aims to explore new biomarkers and to create new medicine.
List of papers published in 2018
Journal
1. Miura T, Matsumoto Y, Kawaguchi T, Masuda Y, Okizaki A, Koga H, Tagami K, Watanabe YS, Uehara Y, Yamaguchi T, Morita T. Low Phase Angle Is Correlated With Worse General Condition in Patients with Advanced Cancer. Nutr Cancer, 71:83-88, 2019
2. Miyashita T, Omori T, Nakamura H, Sugano M, Neri S, Fujii S, Hashimoto H, Tsuboi M, Ochiai A, Ishii G. Spatiotemporal characteristics of fibroblasts-dependent cancer cell invasion. J Cancer Res Clin Oncol, 145:373-381, 2019
3. Kawazoe A, Shitara K, Kuboki Y, Bando H, Kojima T, Yoshino T, Ohtsu A, Ochiai A, Togashi Y, Nishikawa H, Doi T, Kuwata T. Clinicopathological features of 22C3 PD-L1 expression with mismatch repair, Epstein-Barr virus status, and cancer genome alterations in metastatic gastric cancer. Gastric Cancer, 22:69- 76, 2019
4. Kuno I, Yoshida H, Kohno T, Ochiai A, Kato T. Endometrial cancer arising after complete remission of uterine malignant lymphoma: A case report and mutation analysis. Gynecol Oncol Rep, 28:50- 53, 2019
5. Yanagihara K, Kubo T, Mihara K, Kuwata T, Ochiai A, Seyama T, Yokozaki H. Development and Biological Analysis of a Novel Orthotopic Peritoneal Dissemination Mouse Model Generated Using a Pancreatic Ductal Adenocarcinoma Cell Line. Pancreas, 48:315-322, 2019
6. Naito T, Mitsunaga S, Miura S, Tatematsu N, Inano T, Mouri T, Tsuji T, Higashiguchi T, Inui A, Okayama T, Yamaguchi T, Morikawa A, Mori N, Takahashi T, Strasser F, Omae K, Mori K, Takayama K. Feasibility of early multimodal interventions for elderly patients with advanced pancreatic and non-small-cell lung cancer. J Cachexia Sarcopenia Muscle, 10:73-83, 2019
7. Okuyama K, Kitajima Y, Egawa N, Kitagawa H, Ito K, Aishima S, Yanagihara K, Tanaka T, Noshiro H. Mieap-induced accumulation of lysosomes within mitochondria (MALM) regulates gastric cancer cell invasion under hypoxia by suppressing reactive oxygen species accumulation. Sci Rep, 9:2822, 2019
8. Umakoshi M, Takahashi S, Itoh G, Kuriyama S, Sasaki Y, Yanagihara K, Yashiro M, Maeda D, Goto A, Tanaka M. Macrophage- mediated transfer of cancer-derived components to stromal cells contributes to establishment of a pro-tumor microenvironment. Oncogene, 38:2162-2176, 2019
9. Kojima M, Chen Y, Ikeda K, Tsukada Y, Takahashi D, Kawano S, Amemiya K, Ito M, Ohki R, Ochiai A. Recommendation of longterm and systemic management according to the risk factors in rectal NETs patients. Sci Rep, 9:2404, 2019
10. Yanagihara K, Kubo T, Iino Y, Mihara K, Morimoto C, Seyama T, Kuwata T, Ochiai A, Yokozaki H. Development and Biological Analysis of a Novcharacterization of a cancer cahexia model employing a rare human duodenal neuroendocrine carcinoma-originating cell line. Oncotarget, 2019
11. Tsuchimochi M, Yamaguchi H, Hayama K, Okada Y, Kawase T, Suzuki T, Tsubokawa N, Wada N, Ochiai A, Fujii S, Fujii H. Imaging of Metastatic Cancer Cells in Sentinel Lymph Nodes using Affibody Probes and Possibility of a Theranostic Approach. Int J Mol Sci, 20:2019
12. Ohmoto A, Suzuki M, Takai E, Rokutan H, Fujiwara Y, Morizane C, Yanagihara K, Shibata T, Yachida S. Establishment of preclinical chemotherapy models for gastroenteropancreatic neuroendocrine carcinoma. Oncotarget, 9:21086-21099, 2018
13. Watanabe YS, Miura T, Okizaki A, Tagami K, Matsumoto Y, Fujimori M, Morita T, Kinoshita H. Comparison of Indicators for Achievement of Pain Control With a Personalized Pain Goal in a Comprehensive Cancer Center. J Pain Symptom Manage, 55:1159-1164, 2018
14. Kako J, Kobayashi M, Kanno Y, Ogawa A, Miura T, Matsumoto Y. The Optimal Cutoff Point for Expressing Revised Edmonton Symptom Assessment System Scores as Binary Data Indicating the Presence or Absence of Symptoms. Am J Hosp Palliat Care, 35:1390-1393, 2018
15. Tagami K, Okizaki A, Miura T, Watanabe YS, Matsumoto Y, Morita T, Fujimori M, Kinoshita H. Breakthrough Cancer Pain Influences General Activities and Pain Management: A Comparison of Patients with and without Breakthrough Cancer Pain. J Palliat Med, 21:1636-1640, 2018
16. Miura T, Amano K, Shirado A, Baba M, Ozawa T, Nakajima N, Suga A, Matsumoto Y, Shimizu M, Shimoyama S, Kuriyama T, Matsuda Y, Iwashita T, Mori I, Kinoshita H. Low Transthyretin Levels Predict Poor Prognosis in Cancer Patients in Palliative Care Settings. Nutr Cancer, 70:1283-1289, 2018
17. Ichikawa T, Aokage K, Sugano M, Miyoshi T, Kojima M, Fujii S, Kuwata T, Ochiai A, Suzuki K, Tsuboi M, Ishii G. The ratio of cancer cells to stroma within the invasive area is a histologic prognostic parameter of lung adenocarcinoma. Lung Cancer, 118:30- 35, 2018
18. Nakasone S, Mimaki S, Ichikawa T, Aokage K, Miyoshi T, Sugano M, Kojima M, Fujii S, Kuwata T, Ochiai A, Tsuboi M, Goto K, Tsuchihara K, Ishii G. Podoplanin-positive cancer-associated fibroblast recruitment within cancer stroma is associated with a higher number of single nucleotide variants in cancer cells in lung adenocarcinoma. J Cancer Res Clin Oncol, 144:893-900, 2018
19. Makinoshima H, Umemura S, Suzuki A, Nakanishi H, Maruyama A, Udagawa H, Mimaki S, Matsumoto S, Niho S, Ishii G, Tsuboi M, Ochiai A, Esumi H, Sasaki T, Goto K, Tsuchihara K. Metabolic Determinants of Sensitivity to Phosphatidylinositol 3-Kinase Pathway Inhibitor in Small-Cell Lung Carcinoma. Cancer Res, 78:2179-2190, 2018
20. Yamazaki S, Higuchi Y, Ishibashi M, Hashimoto H, Yasunaga M, Matsumura Y, Tsuchihara K, Tsuboi M, Goto K, Ochiai A, Ishii G. Collagen type I induces EGFR-TKI resistance in EGFR-mutated cancer cells by mTOR activation through Akt-independent pathway. Cancer Sci, 109:2063-2073, 2018
21. Katsumata S, Aokage K, Miyoshi T, Tane K, Nakamura H, Sugano M, Kojima M, Fujii S, Kuwata T, Ochiai A, Hayashi R, Tsuboi M, Ishii G. Differences of tumor microenvironment between stage I lepidic-positive and lepidic-negative lung adenocarcinomas. J Thorac Cardiovasc Surg, 156:1679-1688.e2, 2018
22. Sakai T, Aokage K, Neri S, Nakamura H, Nomura S, Tane K, Miyoshi T, Sugano M, Kojima M, Fujii S, Kuwata T, Ochiai A, Iyoda A, Tsuboi M, Ishii G. Link between tumor-promoting fibrous microenvironment and an immunosuppressive microenvironment in 446 stage I lung adenocarcinoma. Lung Cancer, 126:64-71, 2018
23. Kuboki Y, Schatz CA, Koechert K, Schubert S, Feng J, Wittemer- Rump S, Ziegelbauer K, Krahn T, Nagatsuma AK, Ochiai A. In situ analysis of FGFR2 mRNA and comparison with FGFR2 gene copy number by dual-color in situ hybridization in a large cohort of gastric cancer patients. Gastric Cancer, 21:401-412, 2018
24. Kumata H, Murakami K, Ishida K, Miyagi S, Arakawa A, Inayama Y, Kinowaki K, Ochiai A, Kojima M, Higashi M, Moritani S, Kuwahara K, Nakatani Y, Kajiura D, Tamura G, Kijima H, Yamakawa M, Shiraishi T, Inadome Y, Murakami K, Suzuki H, Sawai T, Unno M, Kamei T, Sasano H. Steroidogenesis in ovarian-like mesenchymal stroma of hepatic and pancreatic mucinous cystic neoplasms. Hepatol Res, 48:989-999, 2018
25. Okubo S, Mitsunaga S, Kato Y, Kojima M, Sugimoto M, Gotohda N, Takahashi S, Hayashi R, Konishi M. The prognostic impact of differentiation at the invasive front of biliary tract cancer. J Surg Oncol, 117:1278-1287, 2018
26. Takahashi D, Kojima M, Suzuki T, Sugimoto M, Kobayashi S, Takahashi S, Konishi M, Gotohda N, Ikeda M, Nakatsura T, Ochiai A, Nagino M. Profiling the Tumour Immune Microenvironment in Pancreatic Neuroendocrine Neoplasms with Multispectral Imaging Indicates Distinct Subpopulation Characteristics Concordant with WHO 2017 Classification. Sci Rep, 8:13166, 2018
27. Sato Y, Matsuda S, Maruyama A, Nakayama J, Miyashita T, Udagawa H, Umemura S, Yanagihara K, Ochiai A, Tomita M, Soga T, Tsuchihara K, Makinoshima H. Metabolic Characterization of Antifolate Responsiveness and Non-responsiveness in Malignant Pleural Mesothelioma Cells. Front Pharmacol, 9:1129, 2018
28. Terawaki K, Kashiwase Y, Uzu M, Nonaka M, Sawada Y, Miyano K, Higami Y, Yanagihara K, Yamamoto M, Uezono Y. Leukemia inhibitory factor via the Toll-like receptor 5 signaling pathway involves aggravation of cachexia induced by human gastric cancer- derived 85As2 cells in rats. Oncotarget, 9:34748-34764, 2018
29. Kimura R, Yoneshige A, Hagiyama M, Otani T, Inoue T, Shiraishi N, Yanagihara K, Wakayama T, Ito A. Expression of cell adhesion molecule 1 in gastric neck and base glandular cells: Possible involvement in peritoneal dissemination of signet ring cells. Life Sci, 213:206-213, 2018
30. Tajirika T, Tokumaru Y, Taniguchi K, Sugito N, Matsuhashi N, Futamura M, Yanagihara K, Akao Y, Yoshida K. DEAD-Box Protein RNA-Helicase DDX6 Regulates the Expression of HER2 and FGFR2 at the Post-Transcriptional Step in Gastric Cancer Cells. Int J Mol Sci, 19:2018
31. Yokoyama T, Terawaki K, Minami K, Miyano K, Nonaka M, Uzu M, Kashiwase Y, Yanagihara K, Ueta Y, Uezono Y. Modulation of synaptic inputs in magnocellular neurones in a rat model of cancer cachexia. J Neuroendocrinol, 30:e12630, 2018
32. Takei Y, Shen G, Morita-Kondo A, Hara T, Mihara K, Yanagihara K. MicroRNAs Associated with Epithelial-Mesenchymal Transition Can Be Targeted to Inhibit Peritoneal Dissemination of Human Scirrhous Gastric Cancers. Pathobiology, 85:232-246, 2018
33. Saeki N, Saito A, Sugaya Y, Amemiya M, Ono H, Komatsuzaki R, Yanagihara K, Sasaki H. Chromatin Immunoprecipitation and DNA Sequencing Identified a LIMS1/ILK Pathway Regulated by LMO1 in Neuroblastoma. Cancer Genomics Proteomics. 15(3):165-174,2018 May-Jun.
34. Miura T, Mitsunaga S, Ikeda M, Ohno I, Takahashi H, Kuwata T, Ochiai A. Neural Invasion Spreads Macrophage-Related Allodynia via Neural Root in Pancreatic Cancer. Anesth Analg, 126:1729- 1738, 2018
35 Kiyozumi Y, Iwatsuki M, Kurashige J, Ogata Y, Yamashita K, Koga Y, Toihata T, Hiyoshi Y, Ishimoto T, Baba Y, Miyamoto Y, Yoshida N, Yanagihara K, Mimori K, Baba H. PLOD2 as a potential regulator of peritoneal dissemination in gastric cancer. Int J Cancer, 143:1202-1211, 2018
36. Miura T, Mitsunaga S, Ikeda M, Ohno I, Takahashi H, Suzuki H, Irisawa A, Kuwata T, Ochiai A. Characterization of low active ghrelin ratio in patients with advanced pancreatic cancer. Support Care Cancer, 26:3811-3817, 2018
37. Yanagihara K, Kubo T, Mihara K, Kuwata T, Ochiai A, Seyama T, Yokozaki H. Establishment of a novel cell line from a rare human duodenal poorly differentiated neuroendocrine carcinoma. Oncotarget, 9:36503-36514, 2018
38. Ueda T, Aokage K, Mimaki S, Tane K, Miyoshi T, Sugano M, Kojima M, Fujii S, Kuwata T, Ochiai A, Kusumoto M, Suzuki K, Tsuchihara K, Nishikawa H, Goto K, Tsuboi M, Ishii G. Characterization of the tumor immune-microenvironment of lung adenocarcinoma associated with usual interstitial pneumonia. Lung Cancer, 126:162-169, 2018
39. Ueda T, Aokage K, Nishikawa H, Neri S, Nakamura H, Sugano M, Tane K, Miyoshi T, Kojima M, Fujii S, Kuwata T, Ochiai A, Kusumoto M, Suzuki K, Tsuboi M, Ishii G. Immunosuppressive tumor microenvironment of usual interstitial pneumonia-associated squamous cell carcinoma of the lung. J Cancer Res Clin Oncol, 144:835-844, 2018
