Annual Report 2018
Division of Cancer Immunology
Hiroyoshi Nishikawa, Yoshihiro Ohue, Takuma Irie, Chika Sakai, Kota Itahashi, Eri Sugiyama, Sho Watanabe, Yuki Fujioka, Shota Fukuoka, Ryo Ariyasu, Takahiro Kamata, Shogo Kumagai, Tokiyoshi Tanegashima, Ayaka Tsuge, Joji Nagasaki, Miki Yamazaki, Sho Isoyama, Tamiyo Kobayashi, Sayoko Shinya
Introduction
Recent success of cancer immunotherapy, particularly immune checkpoint inhibitors, makes it a key strategy for cancer treatment and a lot of clinical trials are under way to develop new reagents. Among patients receiving cancer immunotherapy, while some experience longlasting clinical benefits, more than half of the patients fail to exhibit any clinical benefits. Moreover, some patients experience recurrence after initially responding to such immunotherapy. It is therefore urgently required to identify predictive biomarkers, and to develop more effective therapies and optimal immunotherapies for individual patients.
The Team and What We Do
In our laboratory, viable immune cells both in tumors and in the periphery are being investigated using several techniques such as multi-color flow-cytometry, CyTOF, and single cell RNA sequencing by collaboration with the clinical department (Figure 1). A realtime immune monitoring system in cancer patients during their cancer treatment has been established, through which we have had collaborations with pharmaceutical companies in their clinical trials. Especially, regulatory T cells (Treg cells), which have a strong immune suppressive function and are known to be a critical component to inhibit anti-tumor immunity, are analyzed with our original assay. Furthermore, with the findings from clinical samples analyses, we also revere-translate them into basic research to elucidate the essential mechanisms of immune responses to cancer.
Figure 1. Our investigation of dynamic immune state in
cancer patients
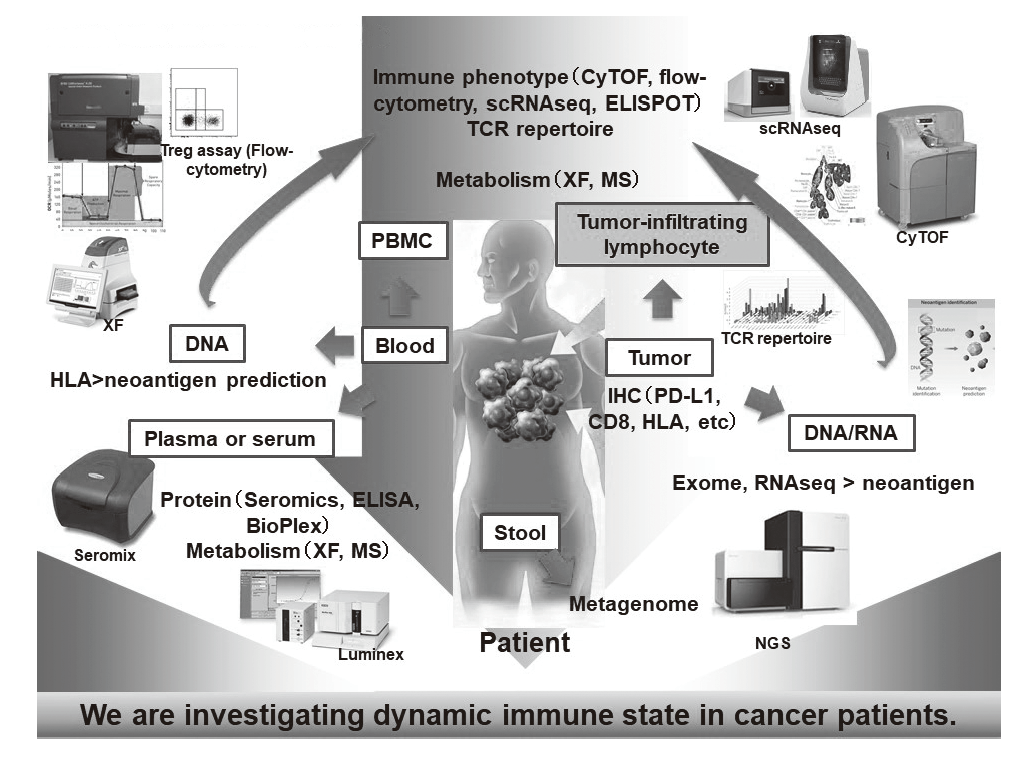

Research activities
When the data from our assays of immune status in a tumor microenvironment were integrated with genomic analyses in non-small cell lung cancer (NSCLC), significant differences between EGFR-mutated and wild-type NSCLC were identified (manuscript is submitted). In addition, we have found a specific driver mutation in gastric cancer which influenced the immune status in a tumor microenvironment (manuscript is submitted).
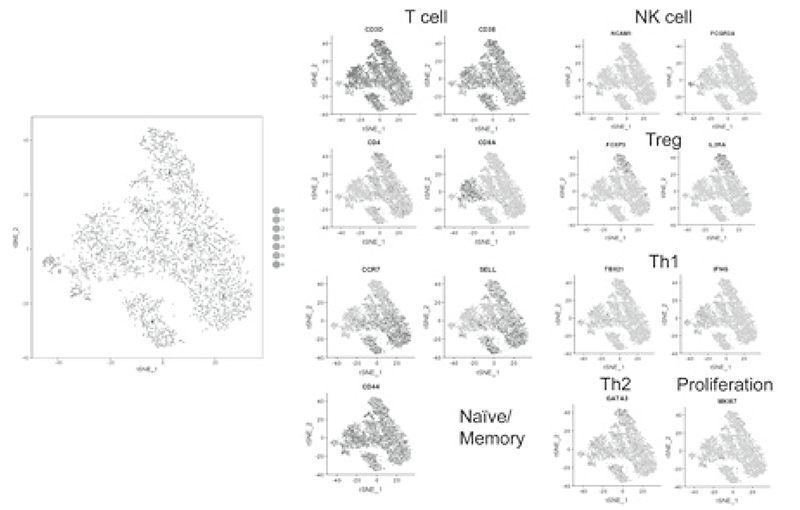
Using our real-time immune monitoring system, we also addressed kinetic changes of immune status in a tumor microenvironment after cancer therapies including not only immune checkpoint inhibitors, but also cytotoxic reagents. We found a reduction of activated Treg cells by a first-in-human clinical reagent using our realtime immune monitoring system, proposing a novel effect of this drug. Furthermore, a specific immune state change in hyperprogression disease (HPD) and a possible predictive biomarker have been identified in patients treated with immune checkpoint inhibitors. We have also worked on the GAPFREE2 project in which tumor-infiltrating lymphocytes at both pre- and post-PD-1 blockade are analyzed using scRNAseq (Figure 2).
Figure 2. Representative data of TILs analyzed by
mass cytometry

Clinical trials
We have collaborated with several pharmaceutical companies in their clinical trials to elucidate predictive biomarkers and immune status in a tumor microenvironment using our real-time immune monitoring system.
Education
Many graduate school students are trained in our Division, and young residents in the National Cancer Center East are also trained to become physician scientists. After finishing their thesis, they continue to study cancer immunology abroad.
Future prospects
ICB has undoubtedly opened a new era in cancer treatment. However, we do not yet know in detail how the human immune system recognizes the tumor. In order to solve this question, immunological analyses using clinical samples are essential.
Therefore, it is important to develop better cancer immunotherapies such as combinations and to identify biomarkers that can stratify responders from non-responders via clarifying the immune suppressive networks in TME
List of papers published in 2018
Journal
1. Watanabe S, Honma Y, Murakami N, Igaki H, Mori T, Hirano H, Okita N, Shoji H, Iwasa S, Takashima A, Kato K, Kobayashi K, Matsumoto F, Yoshimoto S, Itami J, Boku N. Induction chemotherapy with docetaxel, cisplatin and fluorouracil followed by concurrent chemoradiotherapy for unresectable sinonasal undifferentiated carcinoma: Two cases of report. World J Clin Cases, 7:765-772, 2019
2. Kawazoe A, Shitara K, Kuboki Y, Bando H, Kojima T, Yoshino T, Ohtsu A, Ochiai A, Togashi Y, Nishikawa H, Doi T, Kuwata T. Clinicopathological features of 22C3 PD-L1 expression with mismatch repair, Epstein-Barr virus status, and cancer genome alterations in metastatic gastric cancer. Gastric Cancer, 22:69- 76, 2019
3. Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat Rev Clin Oncol, 16:356-371, 2019
4. Inozume T, Yaguchi T, Ariyasu R, Togashi Y, Ohnuma T, Honobe A, Nishikawa H, Kawakami Y, Kawamura T. Analysis of the Tumor Reactivity of Tumor-Infiltrating Lymphocytes in a Metastatic Melanoma Lesion that Lost Major Histocompatibility Complex Class I Expression after Anti-PD-1 Therapy. J Invest Dermatol, 2019
5. Ha D, Tanaka A, Kibayashi T, Tanemura A, Sugiyama D, Wing JB, Lim EL, Teng KWW, Adeegbe D, Newell EW, Katayama I, Nishikawa H, Sakaguchi S. Differential control of human Treg and effector T cells in tumor immunity by Fc-engineered anti-CTLA-4 antibody. Proc Natl Acad Sci USA, 116:609-618, 2019
6. Watanabe S, Hayashi H, Haratani K, Shimizu S, Tanizaki J, Sakai K, Kawakami H, Yonesaka K, Tsurutani J, Togashi Y, Nishio K, Ito A, Nakagawa K. Mutational activation of the epidermal growth factor receptor down-regulates major histocompatibility complex class I expression via the extracellular signal-regulated kinase in non-small cell lung cancer. Cancer Sci, 110:52-60, 2019
7. Itahashi K, Shimizu T, Koyama T, Kondo S, Fujiwara Y, Yamamoto N. Global trends in the distribution of cancer types among patients in oncology phase I trials, 1991-2015. Invest New Drugs, 37:166-174, 2019
8. Kamada T, Togashi Y, Tay C, Ha D, Sasaki A, Nakamura Y, Sato E, Fukuoka S, Tada Y, Tanaka A, Morikawa H, Kawazoe A, Kinoshita T, Shitara K, Sakaguchi S, Nishikawa H. PD-1+ regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc Natl Acad Sci USA, 116:9999-10008, 2019
9. Kochin V, Nishikawa H. Editors' Choice Meddling with meddlers: curbing regulatory T cells and augmenting antitumor immunity. Nagoya J Med Sci, 81:1-18, 2019
10. Shitara K, Nishikawa H. Regulatory T cells: a potential target in cancer immunotherapy. Ann N Y Acad Sci, 1417:104-115, 2018
11. Tada Y, Togashi Y, Kotani D, Kuwata T, Sato E, Kawazoe A, Doi T, Wada H, Nishikawa H, Shitara K. Targeting VEGFR2 with Ramucirumab strongly impacts effector/ activated regulatory T cells and CD8+ T cells in the tumor microenvironment. J Immunother Cancer, 6:106, 2018
12. Takeuchi Y, Tanemura A, Tada Y, Katayama I, Kumanogoh A, Nishikawa H. Clinical response to PD-1 blockade correlates with a sub-fraction of peripheral central memory CD4+ T cells in patients with malignant melanoma. Int Immunol, 30:13-22, 2018
13. Takahashi N, Nishiwaki K, Nakaseko C, Aotsuka N, Sano K, Ohwada C, Kuroki J, Kimura H, Tokuhira M, Mitani K, Fujikawa K, Iwase O, Ohishi K, Kimura F, Fukuda T, Tanosaki S, Takahashi S, Kameoka Y, Nishikawa H, Wakita H. Treatment-free remission after two-year consolidation therapy with nilotinib in patients with chronic myeloid leukemia: STAT2 trial in Japan. Haematologica, 103:1835-1842, 2018
14. Miao W, Sakai K, Ozawa N, Nishiuchi T, Suzuki Y, Ito K, Morioka T, Umitsu M, Takagi J, Suga H, Matsumoto K. Cellular signaling and gene expression profiles evoked by a bivalent macrocyclic peptide that serves as an artificial MET receptor agonist. Sci Rep, 8:16492, 2018
15. Ueda T, Aokage K, Mimaki S, Tane K, Miyoshi T, Sugano M, Kojima M, Fujii S, Kuwata T, Ochiai A, Kusumoto M, Suzuki K, Tsuchihara K, Nishikawa H, Goto K, Tsuboi M, Ishii G. Characterization of the tumor immune-microenvironment of lung adenocarcinoma associated with usual interstitial pneumonia. Lung Cancer, 126:162-169, 2018
16. Itahashi K, Kondo S, Kubo T, Fujiwara Y, Kato M, Ichikawa H, Koyama T, Tokumasu R, Xu J, Huettner CS, Michelini VV, Parida L, Kohno T, Yamamoto N. Evaluating Clinical Genome Sequence Analysis by Watson for Genomics. Front Med, 5:305, 2018
