Annual Report 2018
Division of Cancer Immunology
Hiroyoshi Nishikawa, Yuka Maeda, Yosuke Togashi, Hitomi Nishinakamura, Kentaro Shinohara, Hiroaki Hiramatsu, Etsuko Tanji, Yuki Ishige, Maki Sugaya, Akihito Maki, Yusuke Muto, Nanae Shimoyama
Introduction
Cancer immunotherapies, particularly immune checkpoint inhibitors such as antiCTLA-4 mAb, anti-PD-1 mAb and anti-PD-L1 mAb, are becoming important strategies to treat cancer. However, the clinical effects of immune checkpoint inhibitors are observed in limited patients because cancer cells establish a complex immune suppressive mechanism that inhibits effective anti-tumor immune responses. To reveal the mechanisms is an urgent requirement for the development of effective cancer immunotherapy (Figure 1). Our aims are clarifying the detailed mechanism of immune suppression in a tumor microenvironment by immunologic and genetic approaches and identifying novel approaches to control anti-tumor immune responses.
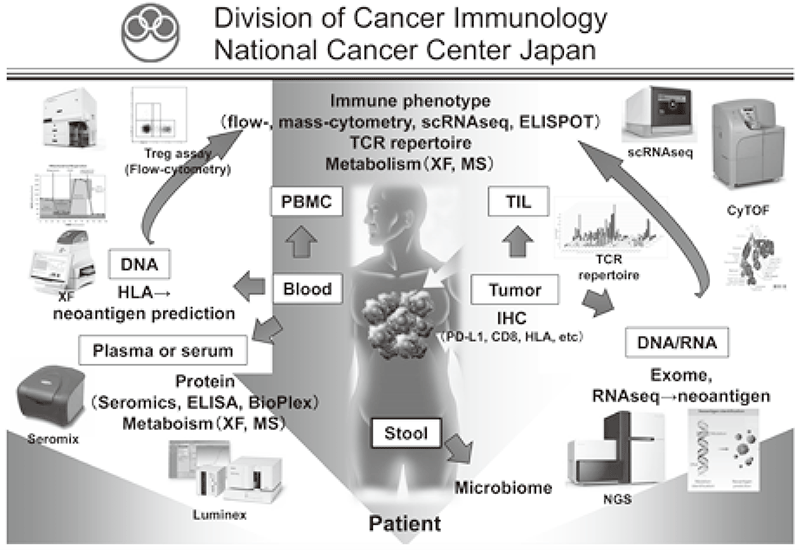
Figure 1. Investigation of dynamic immune state in cancer patients

The Team and What We Do
Our division focused on elucidating the detailed mechanisms involved in tumor immune suppression or tumor immune evasion observed in various types of cancers. We have collected tissues and peripheral blood pre- and posttreatment with immune checkpoint inhibitors from many cancer patients. To comprehensively explore immune responses, molecular markers and gene expression profiles of TILs and PBMCs were examined.
Research activities
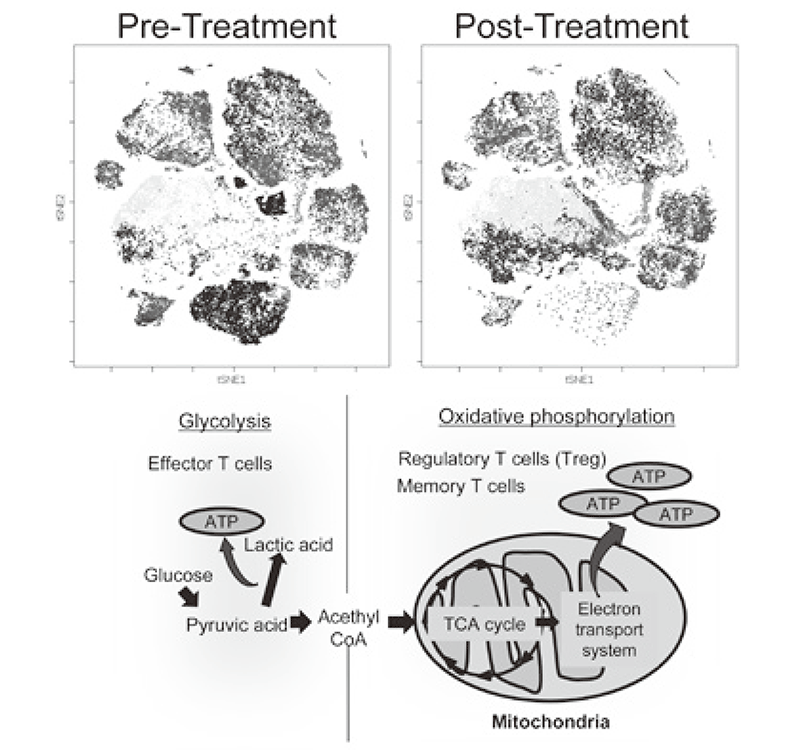
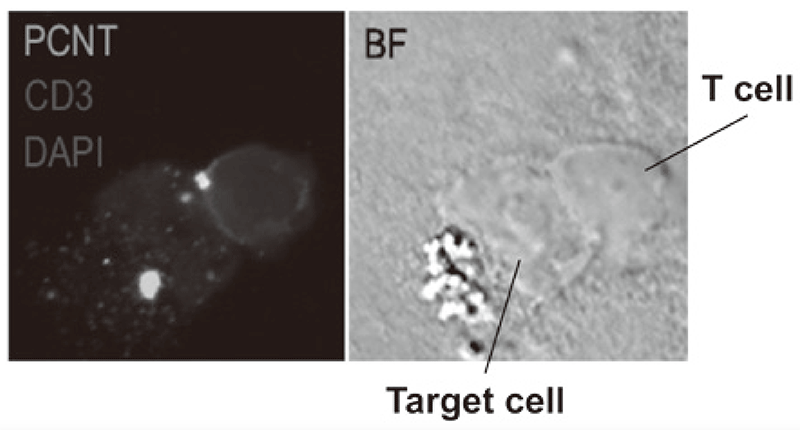
We established strategies to investigate multi-parameters with single cell level using flow-cytometry and mass cytometry (CyTOF). Lymphocytes are known to change their metabolism between glycolysis and oxidative phosphorylation according to their activity states and cell types such as effector T cells and regulatory T cells. We then interrogated the control mechanism of metabolic shift of lymphocytes in immune suppression or immune evasion in a tumor microenvironment (Figure 2). Additionally, to investigate anti-tumor immune responses in vivo, particularly focusing on a tumor microenvironment, we developed mouse models and explored the pattern of the immunological phenotype according to the development/regression of tumors. Further, we also established a new method to evaluate the dynamics of molecular delivery within lymphocytes using laser confocal microscopy.
Figure 2. Representative image of clinical-sample analysis (pre- and post-treatments), and metabolic pathway map in lymphocytes
Clinical trials
We have collaborated with several pharmaceutical companies in their clinical trials to elucidate predictive biomarkers and immune status in a tumor microenvironment using our real-time immune monitoring system.
Figure 3. Confocal microscopical image of molecular dynamics in lymphocytes

Education
Many graduate school students are trained in our Division, and young residents in the National Cancer Center East are also trained to become physician scientists. After finishing their thesis, they continue to study cancer immunology abroad.
Future prospects
Our division aims at elucidating the mechanisms involved in tumor immune suppression or tumor immune evasion in various types of cancers with a broad view such as immunology and genetics. In addition, we plan to investigate immune responses in more detail by integrated analyses using not only immunological methods, but also single-cell based DNA/RNA seq, gene-editing and data science with humans and animal models. We pursue novel effective cancer immunotherapies and identify promising biomarkers based on the insights obtained from our fundamental research.
List of papers published in 2018
Journal
1. Kawazoe A, Shitara K, Kuboki Y, Bando H, Kojima T, Yoshino T, Ohtsu A, Ochiai A, Togashi Y, Nishikawa H, Doi T, Kuwata T. Clinicopathological features of 22C3 PD-L1 expression with mismatch repair, Epstein-Barr virus status, and cancer genome alterations in metastatic gastric cancer. Gastric Cancer, 22:69- 76, 2019
2. Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat Rev Clin Oncol, 16:356-371, 2019
3. Inozume T, Yaguchi T, Ariyasu R, Togashi Y, Ohnuma T, Honobe A, Nishikawa H, Kawakami Y, Kawamura T. Analysis of the Tumor Reactivity of Tumor-Infiltrating Lymphocytes in a Metastatic Melanoma Lesion that Lost Major Histocompatibility Complex Class I Expression after Anti-PD-1 Therapy. J Invest Dermatol, 2019
4. Ha D, Tanaka A, Kibayashi T, Tanemura A, Sugiyama D, Wing JB, Lim EL, Teng KWW, Adeegbe D, Newell EW, Katayama I, Nishikawa H, Sakaguchi S. Differential control of human Treg and effector T cells in tumor immunity by Fc-engineered anti-CTLA-4 antibody. Proc Natl Acad Sci USA, 116:609-618, 2019
5. Watanabe S, Hayashi H, Haratani K, Shimizu S, Tanizaki J, Sakai K, Kawakami H, Yonesaka K, Tsurutani J, Togashi Y, Nishio K, Ito A, Nakagawa K. Mutational activation of the epidermal growth factor receptor down-regulates major histocompatibility complex class I expression via the extracellular signal-regulated kinase in non-small cell lung cancer. Cancer Sci, 110:52-60, 2019
6. Itahashi K, Shimizu T, Koyama T, Kondo S, Fujiwara Y, Yamamoto N. Global trends in the distribution of cancer types among patients in oncology phase I trials, 1991-2015. Invest New Drugs, 37:166-174, 2019
7. Kamada T, Togashi Y, Tay C, Ha D, Sasaki A, Nakamura Y, Sato E, Fukuoka S, Tada Y, Tanaka A, Morikawa H, Kawazoe A, Kinoshita T, Shitara K, Sakaguchi S, Nishikawa H. PD-1+ regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc Natl Acad Sci USA, 116:9999-10008, 2019
8. Kochin V, Nishikawa H. Editors' Choice Meddling with meddlers: curbing regulatory T cells and augmenting antitumor immunity. Nagoya J Med Sci, 81:1-18, 2019
9. Shitara K, Nishikawa H. Regulatory T cells: a potential target in cancer immunotherapy. Ann N Y Acad Sci, 1417:104-115, 2018
10. Tanisawa K, Hirose N, Arai Y, Shimokata H, Yamada Y, Kawai H, Kojima M, Obuchi S, Hirano H, Suzuki H, Fujiwara Y, Taniguchi Y, Shinkai S, Ihara K, Sugaya M, Higuchi M, Arai T, Mori S, Sawabe M, Sato N, Muramatsu M, Tanaka M. Inverse Association Between Height-Increasing Alleles and Extreme Longevity in Japanese Women. J Gerontol A Biol Sci Med Sci, 73:588-595, 2018
11. Tada Y, Togashi Y, Kotani D, Kuwata T, Sato E, Kawazoe A, Doi T, Wada H, Nishikawa H, Shitara K. Targeting VEGFR2 with Ramucirumab strongly impacts effector/ activated regulatory T cells and CD8+ T cells in the tumor microenvironment. J Immunother Cancer, 6:106, 2018
12. Takeuchi Y, Tanemura A, Tada Y, Katayama I, Kumanogoh A, Nishikawa H. Clinical response to PD-1 blockade correlates with a sub-fraction of peripheral central memory CD4+ T cells in patients with malignant melanoma. Int Immunol, 30:13-22, 2018
13. Takahashi N, Nishiwaki K, Nakaseko C, Aotsuka N, Sano K, Ohwada C, Kuroki J, Kimura H, Tokuhira M, Mitani K, Fujikawa K, Iwase O, Ohishi K, Kimura F, Fukuda T, Tanosaki S, Takahashi S, Kameoka Y, Nishikawa H, Wakita H. Treatment-free remission after two-year consolidation therapy with nilotinib in patients with chronic myeloid leukemia: STAT2 trial in Japan. Haematologica, 103:1835-1842, 2018
14. Ueda T, Aokage K, Mimaki S, Tane K, Miyoshi T, Sugano M, Kojima M, Fujii S, Kuwata T, Ochiai A, Kusumoto M, Suzuki K, Tsuchihara K, Nishikawa H, Goto K, Tsuboi M, Ishii G. Characterization of the tumor immune-microenvironment of lung adenocarcinoma associated with usual interstitial pneumonia. Lung Cancer, 126:162-169, 2018
15. Itahashi K, Kondo S, Kubo T, Fujiwara Y, Kato M, Ichikawa H, Koyama T, Tokumasu R, Xu J, Huettner CS, Michelini VV, Parida L, Kohno T, Yamamoto N. Evaluating Clinical Genome Sequence Analysis by Watson for Genomics. Front Med, 5:305, 2018
