Annual Report 2019
Division of Translational Genomics (Tsukiji Campus)
Takashi Kohno, Hitoshi Ichikawa
Introduction
This division aims to facilitate cancer genome medicine by implementing next-generation sequencing (NGS)-based tumor profiling systems.
The Team and What We Do
This division organizes a clinical sequencing team with staff members from the Department of Experimental Therapeutics, Department of Diagnostic Pathology, Department of Laboratory Medicine and others in the National Cancer Center Hospitals and aims to implement NGS-based tumor profiling systems, such as NCC Oncopanel and NCC Oncopanel-Ped.
Research activities
Implementation of NCC Oncopanel clinical sequencing test
Next-generation sequencing (NGS) of tumor DNA (i.e. clinical sequencing) can guide clinical management by providing diagnostic or prognostic data and facilitating the identification of potential treatment regimens, such as molecular-targeted and immune checkpoint blockade therapies. However, tumor-profiling gene panel tests had not been implemented in routine oncological practice by June 2019. By conducting a prospective gene profiling study (TOP-GEAR project; UMIN000011141), the NCC Oncopanel system was approved by PMDA in the SAKIGAKE program of the MHLW (OncoGuide NCC Oncopanel System) as the first tumor-profiling gene panel test in Japan on December 25, 2018 and started to be reimbursed under the national insurance system from June 2019; we have implemented NGS-based gene-panel tests in routine oncological practice in Japan. The NCC Oncopanel test gives us information on somatic alterations of 114 cancer-associated genes and germline mutations in 13 of them, such as BRCA1, BRCA2, MLH1 and MSH2 (Table). Mutations in and clinical data on cancer patients who received the NCC Oncopanel test in Core, Hub, and Liaison Hospitals for Cancer Genomic Medicine are accumulated in the Center for Cancer Genomics and Advanced Therapeutics (C-CAT) in NCC. We have now developed a novel gene panel test, NCC Oncopanel-Ped, to find actionable alterations in pediatric tumors, and its clinical feasibility and utility are being examined in the TOP-GEAR project.
Table. NCC Oncopanel and other gene panel tests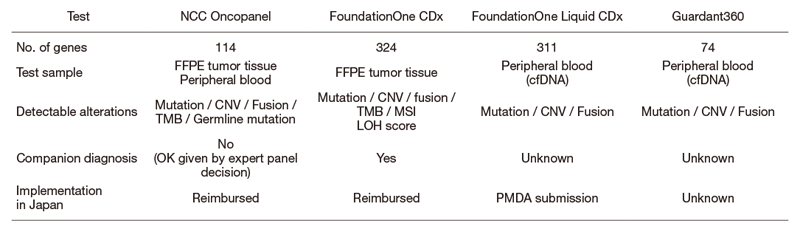

Implementation of clinical sequencing in cancer genome medicine in Japan
In 2018, we validated the feasibility and clinical utility of the NCC Oncopanel test within the Advanced Medical Care B system, including 50 Core or Liaison Hospitals for Cancer Genomic Medicine in Japan. Data analysis of this trial is ongoing.
Clinical trials
TOP-GEAR: Trial of Onco-Panel for Gene-profiling to Estimate both Adverse events and Response by cancer treatment (UMIN000011141)
Education
Post-doctoral fellows and chief residents in NCC were educated through on-the-job training in several translational research projects.
Future prospects
Tumor-profiling multiplex gene panel tests have underpinned cancer genome medicine in Japan. However, a gap remains between the number of patients with actionable mutations and those receiving genomically matched therapy. This gap is largely attributable to the lack of availability/accessibility of relevant trials and drugs. To fill it, we are collaborating with oncologists for the following three purposes: 1) To facilitate molecularly driven clinical trials by implementing tumor genome profiling tests, 2) To annotate VUSs (variants of unknown significance mutations) in druggable genes and 3) To establish clinical trials for gene alterations with a status changing from “undruggable” to “druggable”, such as deleterious mutations in chromatin regulator genes.
List of papers published in 2019
Journal
1. Sunami K, Ichikawa H, Kubo T, Kato M, Fujiwara Y, Shimomura A, Koyama T, Kakishima H, Kitami M, Matsushita H, Furukawa E, Narushima D, Nagai M, Taniguchi H, Motoi N, Sekine S, Maeshima A, Mori T, Watanabe R, Yoshida M, Yoshida A, Yoshida H, Satomi K, Sukeda A, Hashimoto T, Shimizu T, Iwasa S, Yonemori K, Kato K, Morizane C, Ogawa C, Tanabe N, Sugano K, Hiraoka N, Tamura K, Yoshida T, Fujiwara Y, Ochiai A, Yamamoto N, Kohno T. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: A hospital-based study. Cancer Sci, 110:1480-1490, 2019
2. Ikemura S, Yasuda H, Matsumoto S, Kamada M, Hamamoto J, Masuzawa K, Kobayashi K, Manabe T, Arai D, Nakachi I, Kawada I, Ishioka K, Nakamura M, Namkoong H, Naoki K, Ono F, Araki M, Kanada R, Ma B, Hayashi Y, Mimaki S, Yoh K, Kobayashi SS, Kohno T, Okuno Y, Goto K, Tsuchihara K, Soejima K. Molecular dynamics simulation-guided drug sensitivity prediction for lung cancer with rare EGFR mutations. Proc Natl Acad Sci U S A, 116:10025-10030, 2019
3. Kuroda T, Ogiwara H, Sasaki M, Takahashi K, Yoshida H, Kiyokawa T, Sudo K, Tamura K, Kato T, Okamoto A, Kohno T. Therapeutic preferability of gemcitabine for ARID1A-deficient ovarian clear cell carcinoma. Gynecol Oncol, 155:489-498, 2019
4. Wirth LJ, Kohno T, Udagawa H, Ishii G, Ebata KB, Tuch B, Zhu EY, Nguyen M, Smith S, Hanson LM, Burkhard MR, Cable L, Blake JF, Condroski KR, Brandhuber BJ, Andrews S, Rothenberg SM, Goto K. Emergence and targeting of acquired and hereditary resistance to multikinase RET inhibition in RET-altered cancer patients. JCO Prec Oncol, 3:DOI:10.1200/PO.19.00189, 2019
5. Mizuno T, Fujiwara Y, Yoshida K, Kohno T, Ohe Y. Next-Generation Sequencer Analysis of Pulmonary Pleomorphic Carcinoma With a CD74-ROS1 Fusion Successfully Treated With Crizotinib. J Thorac Oncol, 14:e106-e108, 2019
6. Kuno I, Yoshida H, Kohno T, Ochiai A, Kato T. Endometrial cancer arising after complete remission of uterine malignant lymphoma: A case report and mutation analysis. Gynecologic oncology reports, 28:50-53, 2019
7. Kurozumi K, Nakano Y, Ishida J, Tanaka T, Doi M, Hirato J, Yoshida A, Washio K, Shimada A, Kohno T, Ichimura K, Yanai H, Date I. High-grade glioneuronal tumor with an ARHGEF2-NTRK1 fusion gene. Brain Tumor Pathol, 36:121-128, 2019
8. Suzuki A, Onodera K, Matsui K, Seki M, Esumi H, Soga T, Sugano S, Kohno T, Suzuki Y, Tsuchihara K. Characterization of cancer omics and drug perturbations in panels of lung cancer cells. Sci Rep, 9:19529, 2019
9. Nakano Y, Tomiyama A, Kohno T, Yoshida A, Yamasaki K, Ozawa T, Fukuoka K, Fukushima H, Inoue T, Hara J, Sakamoto H, Ichimura K. Identification of a novel KLC1-ROS1 fusion in a case of pediatric low-grade localized glioma. Brain Tumor Pathol, 36:14-19, 2019
10. Seki M, Katsumata E, Suzuki A, Sereewattanawoot S, Sakamoto Y, Mizushima-Sugano J, Sugano S, Kohno T, Frith MC, Tsuchihara K, Suzuki Y. Evaluation and application of RNA-Seq by MinION. DNA Res, 26:55-65, 2019
11. Tsurubuchi T, Nakano Y, Hirato J, Yoshida A, Muroi A, Sakamoto N, Alexander Z, Matsuda M, Ishikawa E, Kohno T, Yoshioka T, Honda-Kitahara M, Ichimura K, Yamamoto T, Matsumura A. Subependymal giant cell astrocytoma harboring a PRRC2B-ALK fusion: A case report. Pediatr Blood Cancer, 66:e27995, 2019
