Annual Report 2019
Division of Epigenomics
Toshikazu Ushijima, Satoshi Yamashita, Hideyuki Takeshima, Naoko Hattori, Harumi Yamada, Takahiro Ebata, Sho Ueda, Mika Wakabayashi, Kazuhiro Nishiyama, Liu Yuyu, Chihiro Takeuchi, Aya Nakajima, Yuko Miyaji, Naoko Takagi, Kana Hashimoto, Chun-Dong Zhang, Reiko Nagano
Introduction
This Division has been focusing on the epigenetic mechanisms of carcinogenesis, and has identified many aberrantly methylated genes in various cancers, including gastric cancers, esophageal squamous cell carcinomas (ESCCs), neuroblastomas, breast cancers, pancreatic cancers, lung cancers, ovarian cancers, and melanomas. These findings have led to identification of novel tumor-suppressor genes, development of powerful biomarkers, and establishment of the concept of an “epigenetic field for cancerization (epigenetic field defect)”. This Division continues its activity in 1) revealing induction mechanisms of epigenetic alterations, 2) developing clinically useful biomarkers, and 3) developing an epigenetic cancer therapy.
Research activities
1. Elucidation of Induction Mechanisms of Epigenetic Alterations
Revealing induction mechanisms of epigenetic alterations is very important in understanding cancers. This year, it was revealed that inflammation-induced aberrant methylation is not a simple acceleration of age-related methylation, and methylation resistant genes, such as tumor-suppressor genes, could be also methylated by inflammation.
It is possible that epigenetic alterations are also accumulated in a cancer microenvironment, and this might enhance cancer development and progression. This year, by analyzing cancer-associated fibroblasts (CAFs) derived from tissues of gastric cancer patients, it was revealed that various secreted factors that can potentially enhance the malignancy of cancer cells were up-regulated by the epigenome alteration, namely trimethylation of histone H3 lysine 27 (H3K27me3) (Figure 1).
Figure 1.Epigenome alterations induce overexpression of various secreted factors in CAFs.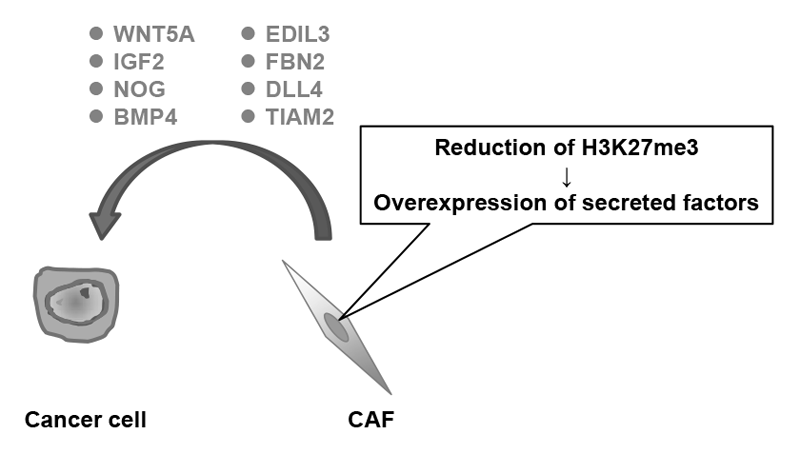

2. Development of Biomarkers
This Division previously has conducted a multicenter prospective cohort study for the prediction of metachronous gastric cancer risk after endoscopic resection, and shown that “the measurement of aberrant DNA methylation accumulated in normal tissues is clinically useful for cancer risk diagnosis”. Based on this result, we are now conducting a multicenter prospective cohort study for the prediction of gastric cancer risk in healthy volunteers who underwent eradication of Helicobacter pylori, the almost exclusive cause of gastric cancers. This year, recruitment of 1,880 people was completed, and we are now conducting follow-up to detect cancer development.
It was also shown that esophageal squamous cell carcinoma (ESCC) patients with methylated FGF5 showed susceptibility to definitive chemoradiotherapy (dCRT).
3. Development of Epigenetic Cancer Therapy
As a strategy to overcome resistance to anticancer drugs or molecular target drugs, a combination therapy using an epigenetic drug is promising. This year, it was revealed that epigenetic priming using a DNA demethylating drug, decitabine, could sensitize gastric cancer cells to irinotecan and cisplatin by restoring multiple pathways.
We also developed two novel prodrugs of decitabine, OR-2003 and OR-2100, that exhibited high stability to cytidine deaminase, comparable DNA demethylation activity, and less toxicity.
Education
We have accepted four trainees from Tokyo Women’s Medical University, Kyorin University, Oita University, and Shonan Kamakura General Hospital.
Future prospects
Based on these results, this Division will 1) continue multicenter prospective cohort studies for the prediction of gastric cancer risk, and 2) conduct the development of epigenetic cancer prevention and therapy.
List of papers published in 2019
Journal
1. Maeda M, Takeshima H, Iida N, Hattori N, Yamashita S, Moro H, Yasukawa Y, Nishiyama K, Hashimoto T, Sekine S, Ishii G, Ochiai A, Fukagawa T, Katai H, Sakai Y, Ushijima T. Cancer cell niche factors secreted from cancer-associated fibroblast by loss of H3K27me3. Gut, 69:243-251, 2020
2. Moro H, Hattori N, Nakamura Y, Kimura K, Imai T, Maeda M, Yashiro M, Ushijima T. Epigenetic priming sensitizes gastric cancer cells to irinotecan and cisplatin by restoring multiple pathways. Gastric Cancer, 23:105-115, 2020
3. Suzuki T, Fukuhara S, Nomoto J, Yamashita S, Maeshima AM, Ito Y, Hatta S, Yuda S, Makita S, Munakata W, Suzuki T, Maruyama D, Taniguchi H, Ushijima T, Izutsu K, Tobinai K, Kobayashi Y. Clinicopathological and Genetic Features of Limited-Stage Diffuse Large B-cell Lymphoma With Late Relapse: Targeted Sequencing Analysis of Gene Alterations in the Initial and Late Relapsed Tumors. Haematologica, 2020
4. Sugimoto K, Ito T, Hulbert A, Chen C, Orita H, Maeda M, Moro H, Fukagawa T, Ushijima T, Katai H, Wada R, Sato K, Sakamoto K, Yu W, Considine M, Cope L, Brock MV. DNA methylation genome-wide analysis in remnant and primary gastric cancers. Gastric Cancer, 22:1109-1120, 2019
5. Hironaka-Mitsuhashi A, Tsuda H, Yoshida M, Shimizu C, Asaga S, Hojo T, Tamura K, Kinoshita T, Ushijima T, Hiraoka N, Fujiwara Y. Invasive breast cancers in adolescent and young adult women show more aggressive immunohistochemical and clinical features than those in women aged 40-44 years. Breast Cancer, 26:386-396, 2019
6. Zong L, Hattori N, Yasukawa Y, Kimura K, Mori A, Seto Y, Ushijima T. LINC00162 confers sensitivity to 5-Aza-2'-deoxycytidine via modulation of an RNA splicing protein, HNRNPH1. Oncogene, 38:5281-5293, 2019
7. Kuzumaki N, Suda Y, Iwasawa C, Narita M, Sone T, Watanabe M, Maekawa A, Matsumoto T, Akamatsu W, Igarashi K, Tamura H, Takeshima H, Tawfik VL, Ushijima T, Hattori N, Okano H, Narita M. Cell-specific overexpression of COMT in dopaminergic neurons of Parkinson's disease. Brain, 142:1675-1689, 2019
8. Zwergel C, Schnekenburger M, Sarno F, Battistelli C, Manara MC, Stazi G, Mazzone R, Fioravanti R, Gros C, Ausseil F, Florean C, Nebbioso A, Strippoli R, Ushijima T, Scotlandi K, Tripodi M, Arimondo PB, Altucci L, Diederich M, Mai A, Valente S. Identification of a novel quinoline-based DNA demethylating compound highly potent in cancer cells. Clin Epigenetics, 11:68, 2019
9. Yatsuoka W, Ueno T, Miyano K, Uezono Y, Enomoto A, Kaneko M, Ota S, Soga T, Sugimoto M, Ushijima T. Metabolomic profiling reveals salivary hypotaurine as a potential early detection marker for medication-related osteonecrosis of the jaw. PLoS One, 14:e0220712, 2019
10. Hattori N, Sako M, Kimura K, Iida N, Takeshima H, Nakata Y, Kono Y, Ushijima T. Novel prodrugs of decitabine with greater metabolic stability and less toxicity. Clin Epigenetics, 11:111, 2019
11. Iwabu J, Yamashita S, Takeshima H, Kishino T, Takahashi T, Oda I, Koyanagi K, Igaki H, Tachimori Y, Daiko H, Nakazato H, Nishiyama K, Lee YC, Hanazaki K, Ushijima T. FGF5 methylation is a sensitivity marker of esophageal squamous cell carcinoma to definitive chemoradiotherapy. Sci Rep, 9:13347, 2019
12. Nakano Y, Hasegawa D, Stewart DR, Schultz KAP, Harris AK, Hirato J, Uemura S, Tamura A, Saito A, Kawamura A, Yoshida M, Yamasaki K, Yamashita S, Ushijima T, Kosaka Y, Ichimura K, Dehner LP, Hill DA. Presacral malignant teratoid neoplasm in association with pathogenic DICER1 variation. Mod Pathol, 32:1744-1750, 2019
13. Pang K, Park J, Ahn SG, Lee J, Park Y, Ooshima A, Mizuno S, Yamashita S, Park KS, Lee SY, Jeong J, Ushijima T, Yang KM, Kim SJ. RNF208, an estrogen-inducible E3 ligase, targets soluble Vimentin to suppress metastasis in triple-negative breast cancers. Nat Commun, 10:5805, 2019
14. Asano N, Takeshima H, Yamashita S, Takamatsu H, Hattori N, Kubo T, Yoshida A, Kobayashi E, Nakayama R, Matsumoto M, Nakamura M, Ichikawa H, Kawai A, Kondo T, Ushijima T. Epigenetic reprogramming underlies efficacy of DNA demethylation therapy in osteosarcomas. Sci Rep, 9:20360, 2019
15. Yamashita S, Nanjo S, Rehnberg E, Iida N, Takeshima H, Ando T, Maekita T, Sugiyama T, Ushijima T. Distinct DNA methylation targets by aging and chronic inflammation: a pilot study using gastric mucosa infected with Helicobacter pylori. Clin Epigenetics, 11:191, 2019
