Annual Report 2020
Division of Epigenomics
Toshikazu Ushijima, Satoshi Yamashita, Hideyuki Takeshima, Naoko Hattori, Harumi Yamada, Takahiro Ebata, Sho Ueda, Kazuhiro Nishiyama, Yuyu Liu, Chihiro Takeuchi, Chun-Dong Zhang, Yoshimi Yasukawa, Masahide Fukuda, Mika Wakabayashi, Kana Hashimoto, Aya Nakajima, Yuko Miyaji, Naoko Takagi
Introduction
This division has been focusing on the epigenetic mechanisms of carcinogenesis, and has identified many aberrantly methylated genes in various cancers, including gastric cancers, esophageal squamous cell carcinomas (ESCCs), neuroblastomas, breast cancers, pancreatic cancers, lung cancers, ovarian cancers, and melanomas. These findings have led to the identification of novel tumor-suppressor genes, development of powerful biomarkers, and establishment of the concept of an "epigenetic field for cancerization (epigenetic field defect)". This division continues its activity in 1) revealing the induction mechanisms of epigenetic alterations, 2) developing clinically useful biomarkers, and 3) developing an epigenetic cancer therapy.
Research activities
1. Elucidation of Induction Mechanisms of Epigenetic Alterations
Revealing the induction mechanisms of epigenetic alterations is very important to understand cancers. This year, expression of TET genes was revealed to down-regulate in inflamed tissues by up-regulation of multiple miRNAs due to NF-κB signaling pathway activation. A vicious combination of TET repression, due to NF-κB activation, and enhancement of DNMT activity, due to nitric oxide exposure, was responsible for aberrant methylation induction by chronic inflammation (Fig. 1).
Figure1. Molecular mechanism of aberrant DNA methylation induction by chronic inflammation 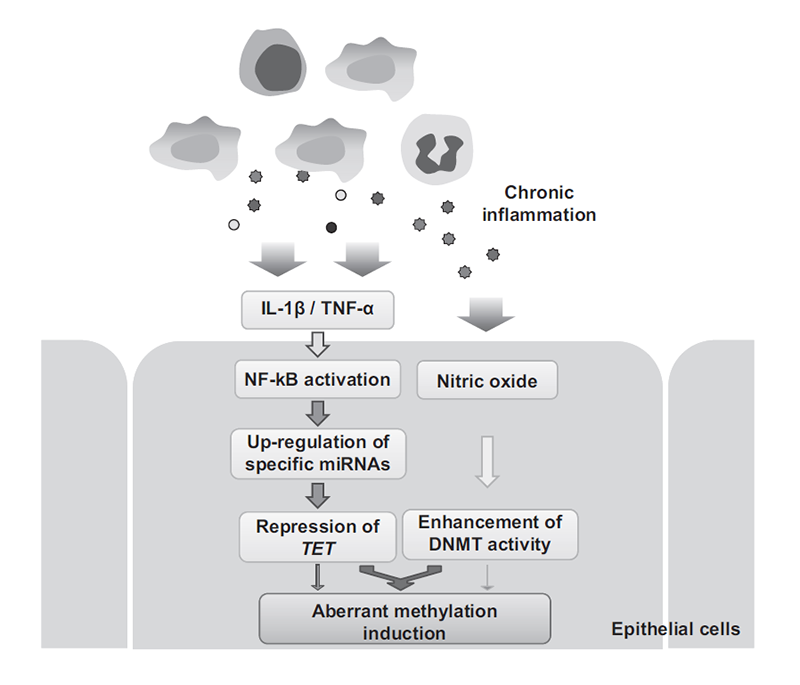

Epigenetic alterations are also accumulated in a cancer microenvironment, and this might enhance cancer development and progression. This year, by analyzing cancer-associated fibroblasts (CAFs) derived from tissues of gastric cancer patients, it was revealed that up-regulation of SAA1 by enhancer activation was important for gastric cancer progression.
2. Development of Biomarkers
This division previously has conducted a multicenter prospective cohort study for predicting metachronous gastric cancer risk after endoscopic resection, and shown that "the measurement of aberrant DNA methylation accumulated in normal tissues is clinically useful for cancer risk diagnosis". Based on this result, a multicenter prospective cohort study is currently being conducted for predicting gastric cancer risk in healthy volunteers who underwent eradication of Helicobacter pylori, the almost exclusive cause of gastric cancers. A follow-up on 1,880 recruited people is currently conducted for detecting cancer development.
It was also shown that even in breast cancer patients without BRCA1 mutations, its promoter methylation was associated with the response to olaparib/eribulin combination therapy.
3. Development of epigenetic cancer therapy
DNA demethylating therapy with low-dose prolonged drug exposure shows a remarkable therapeutic efficacy, despite its small demethylating effect. This year, epigenetic reprogramming of different cancer-related pathways at the single-cell level was revealed to likely underlie the remarkable efficacy of low-dose DNA demethylating therapy.
It was also shown that a novel prodrug of decitabine, OR-2100, collaboratively developed with a pharmaceutical company, was potentially useful for the treatment of ATL.
Education
We have accepted seven trainees from Tokyo Women’s Medical University, Kyorin University, Oita University, Shonan Kamakura General Hospital, University of Tokyo, and Yokohama City University.
Future Prospects
Based on these results, this division will 1) continue to conduct multicenter prospective cohort studies for predicting gastric cancer risk, and 2) conduct the development of epigenetic cancer prevention and therapy.
List of papers published in 2020
Journal
1. Chiang TH, Maeda M, Yamada H, Chan CC, Chen SLS, Chiu SYH, Chen YN, Chou YH, Shieh CF, Liu CY, Chiu HM, Chiang H, Shun CT, Lin MW, Wu MS, Lin JT, Hsiu-Hsi C, Ushijima T, Graham DY, Lee YC. Risk Stratification for Gastric Cancer after Helicobacter pylori Eradication: A Population-based Study on Matsu Islands. J Gastroenterol Hepatol, 36:671-679, 2021
2. Yasukawa Y, Hattori N, Iida N, Takeshima H, Maeda M, Kiyono T, Sekine S, Seto Y, Ushijima T. SAA1 is upregulated in gastric cancer-associated fibroblasts possibly by its enhancer activation. Carcinogenesis, 42:180-189, 2021
3. Sekine S, Yamashita S, Yamada M, Hashimoto T, Ogawa R, Yoshida H, Taniguchi H, Kojima M, Ushijima T, Saito Y. Clinicopathological and molecular correlations in traditional serrated adenoma. J Gastroenterol, 55:418-427, 2020
4. Nakatani Y, Kato K, Shoji H, Iwasa S, Honma Y, Takashima A, Ushijima T, Ito Y, Itami J, Boku N. Comparison of involved field radiotherapy and elective nodal irradiation in combination with concurrent chemotherapy for T1bN0M0 esophageal cancer. Int J Clin Oncol, 25:1098-1104, 2020
5. Kawachi A, Yamashita S, Okochi-Takada E, Hirakawa A, Tsuda H, Shimomura A, Kojima Y, Yonemori K, Fujiwara Y, Kinoshita T, Ushijima T, Tamura K. BRCA1 promoter methylation in breast cancer patients is associated with response to olaparib/eribulin combination therapy. Breast Cancer Res Treat, 181:323-329, 2020
6. Watanabe T, Yamashita S, Ureshino H, Kamachi K, Kurahashi Y, Fukuda-Kurahashi Y, Yoshida N, Hattori N, Nakamura H, Sato A, Kawaguchi A, Sueoka-Aragane N, Kojima K, Okada S, Ushijima T, Kimura S, Sueoka E. Targeting aberrant DNA hypermethylation as a driver of ATL leukemogenesis using the new oral demethylating agent OR-2100. Blood, 136:871-884, 2020
7. Takeshima H, Niwa T, Yamashita S, Takamura-Enya T, Iida N, Wakabayashi M, Nanjo S, Abe M, Sugiyama T, Kim YJ, Ushijima T. TET repression and increased DNMT activity synergistically induce aberrant DNA methylation. J Clin Invest, 130:5370-5379, 2020
8. Blair VR, McLeod M, Carneiro F, Coit DG, D'Addario JL, van Dieren JM, Harris KL, Hoogerbrugge N, Oliveira C, van der Post RS, Arnold J, Benusiglio PR, Bisseling TM, Boussioutas A, Cats A, Charlton A, Schreiber KEC, Davis JL, Pietro M, Fitzgerald RC, Ford JM, Gamet K, Gullo I, Hardwick RH, Huntsman DG, Kaurah P, Kupfer SS, Latchford A, Mansfield PF, Nakajima T, Parry S, Rossaak J, Sugimura H, Svrcek M, Tischkowitz M, Ushijima T, Yamada H, Yang HK, Claydon A, Figueiredo J, Paringatai K, Seruca R, Bougen-Zhukov N, Brew T, Busija S, Carneiro P, DeGregorio L, Fisher H, Gardner E, Godwin TD, Holm KN, Humar B, Lintott CJ, Monroe EC, Muller MD, Norero E, Nouri Y, Paredes J, Sanches JM, Schulpen E, Ribeiro AS, Sporle A, Whitworth J, Zhang L, Reeve AE, Guilford P. Hereditary diffuse gastric cancer: updated clinical practice guidelines. Lancet Oncol, 21:e386-e397, 2020
9. Ishihara H, Yamashita S, Liu YY, Hattori N, El-Omar O, Ikeda T, Fukuda H, Yoshida K, Takagi T, Taneda S, Kondo T, Nagashima Y, Tanabe K, Ushijima T. Genetic and epigenetic profiling indicates the proximal tubule origin of renal cancers in end-stage renal disease. Cancer Sci, 111:4276-4287, 2020
10. Takeshima H, Yoda Y, Wakabayashi M, Hattori N, Yamashita S, Ushijima T. Low-dose DNA demethylating therapy induces reprogramming of diverse cancer-related pathways at the single-cell level. Clin Epigenetics, 12:142, 2020
11. Yamashita S, Hattori N, Fujii S, Yamaguchi T, Takahashi M, Hozumi Y, Kogawa T, El-Omar O, Liu YY, Arai N, Mori A, Higashimoto H, Ushijima T, Mukai H. Multi-omics analyses identify HSD17B4 methylation-silencing as a predictive and response marker of HER2-positive breast cancer to HER2-directed therapy. Sci Rep, 10:15530, 2020
12. Mukai H, Yamaguchi T, Takahashi M, Hozumi Y, Fujisawa T, Ohsumi S, Akabane H, Nishimura R, Takashima T, Park Y, Sagara Y, Toyama T, Imoto S, Mizuno T, Yamashita S, Fujii S, Uemura Y. Ki-67 response-guided preoperative chemotherapy for HER2-positive breast cancer: results of a randomised Phase 2 study. Br J Cancer, 122:1747-1753, 2020
