Annual Report 2022
Division of Cancer Immunology
Hiroyoshi Nishikawa, Shohei Koyama, Takuma Irie, Kota Itahashi, Kosuke Tanaka, Yasuko Tada, Shogo Kumagai, Genki Okumura, Sho Watanabe, Rika Fujii, Akihito Kawazoe, Junichiro Yuda, Megumi Fukuoka, Yi-tzu Lin, Atsuo Sai, Sota Nakagawa, Daisuke Ito, Takuya Owari, Go Watanabe, Masaki Kondo, Tomohiro Iwasawa, Fumihiro Terasaki, Kazuhiro Kumagai, Tomoya Suzuki, Tamiyo Kobayashi, Sho Isoyama, Konomi Onagawa, Tomoka Takaku, Megumi Takemura, Megumi Hoshino, Chie Ozawa, Mizuho Kikuchi, Yoko Ohira, Sayuri Yoshimatsu
Introduction
Cancer immunotherapy, represented by immune checkpoint inhibitors (ICIs), has become one of the standard treatment options for various types of cancer. These ICIs are frequently administered in combination with conventional drugs in clinical settings. Various biomarkers such as PD-L1 expression in tumors and tumor mutation burden (TMB) have been identified to predict therapeutic responses, demonstrating their effectiveness in specific cancers. However, our understanding of markers based on stratification by immuno-genomic analysis of the tumor microenvironment (TME) that can accurately predict the therapeutic efficacy of ICIs remains limited. In addition, as the utilization of ICI therapy expands, the number of cases of long-term response and relapse after long-term treatment is increasing, and the mechanism(s) for long-term response and the resistance to ICIs needs to be clarified.
The Team and What We Do
To address these challenges, our laboratory collaborate with clinical teams to identify novel biomarkers related to clinical responses by ICIs, to elucidate the mechanisms involved in treatment resistance to ICIs, and to propose combination treatment strategies targeting these mechanisms. Focusing on regulatory T cells (Tregs), which play a major role in suppressing anti-tumor immunity, we analyze tumor tissues, blood, and stools before and after various treatments including ICIs, and comprehensively analyze both cancer cells and immune cells by multicolor flow cytometry, multiplex immunohistochemistry, CyTOF, single cell RNA/ATAC/TCR seq, whole exome analysis, and metagenomic analysis (Figure 1). In addition, we investigate the mechanisms of the phenomena observed in humans by transferring to mouse models. We are also actively involved in collaborative research with pharmaceutical companies, aiming at introducing new biomarkers into clinical trials and developing new immunotherapies and combination cancer therapies.
Figure 1. We are investigating the dynamic immune state in cancer patients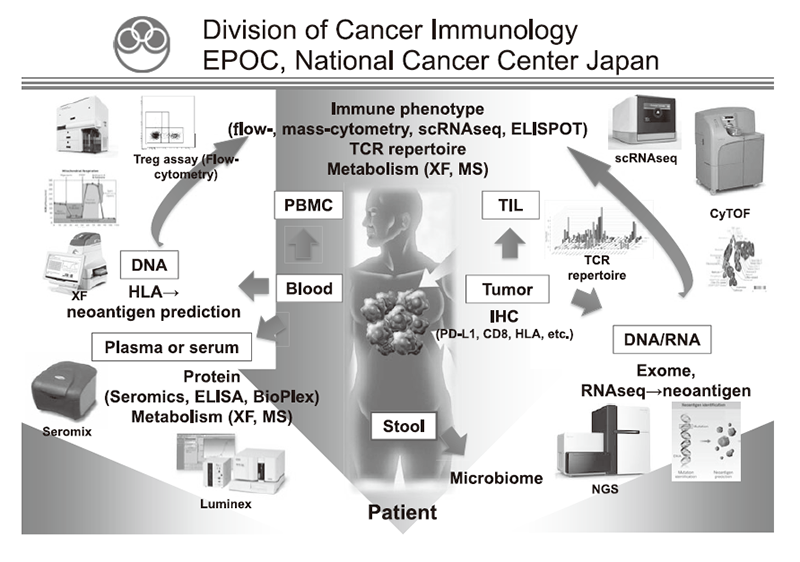

Figure 2. Developing immuno-precision biomarkers and elucidating the mechanisms of PD-1 expression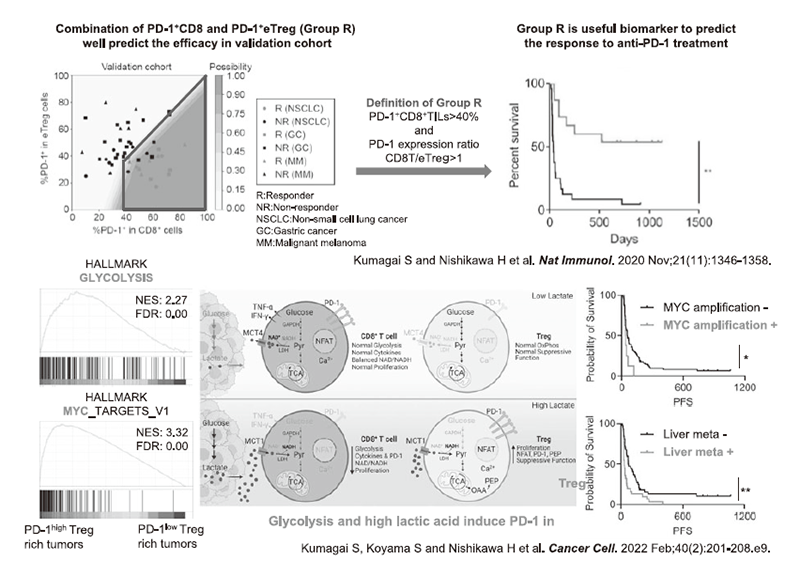

Figure 3. Elucidating Treg cell diversity and differentiation in the TME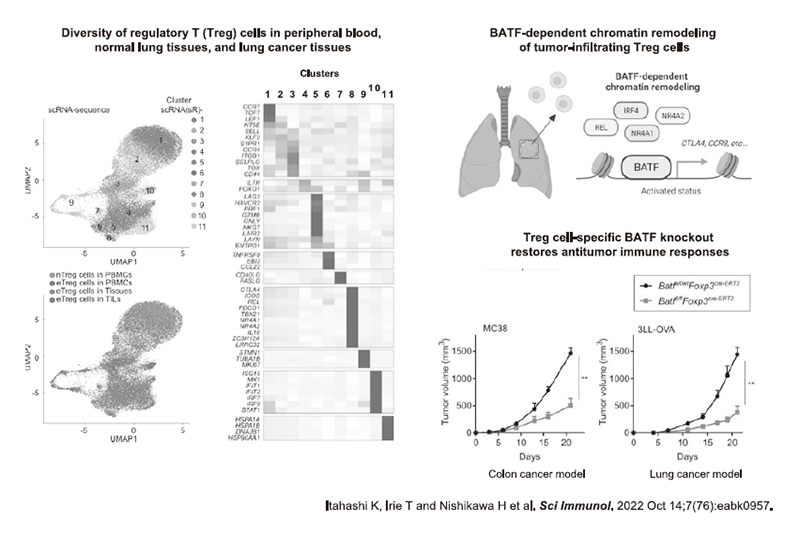

Research Activities
In collaboration with a clinical team led by the Department of Gastroenterology and Respiratory Medicine in NCCE and NCCH, our laboratory conducted a research on biomarkers for predicting the therapeutic responses to anti-PD-1 antibodies, and identified that the balance of PD-1 expression between CD8+ T cells and Tregs is crucial to stratify responders from non-responders based on analysis of tumor-infiltrating T cells (Kumagai S et al. Nat Immunol 2020). However, the factors that regulate PD-1 expression in Tregs were unknown. Detailed analysis revealed that in tumors where the glycolysis is activated (e.g., MYC amplification and metastatic liver tumors), lactate production is enhanced and can be utilized as an energy source only by Tregs, but not by CD8+ T cells, which induces PD-1 expression in Treg compared to CD8+ T cells (Figure 2) (Kumagai S et al. Cancer Cell 2022). Furthermore, in collaboration with the Department of Respiratory Surgery and Clinical Pathology, we examined the transcription factors that regulate the activation of Tregs during their invasion from the peripheral blood into tumor tissues at the epigenomic and gene expression levels to develop Treg-targeted therapy, and found that the transcription factor BATF is essential for regulating the suppressive activity of Tregs in the TME (Figure 3) (Itahashi K et al. Sci Immunol 2022).
Clinical Trials
We have collaborated with several pharmaceutical companies in their clinical trials to elucidate biomarkers for predicting therapeutic responses and to evaluate immune status in the TME using our real-time immune monitoring.
Education
Many graduate school students are trained in our division, and young residents in NCCE are also trained to become physician scientists. After finishing their PhD course, some the students continue their cancer immunology studies abroad.
Future Prospects
ICIs have undoubtedly opened a new era in cancer treatment. However, there are still several issues to be clarified, including acquired resistance and suppressive immune modulation by somatic mutations in cancer cells, for overcoming the drawbacks of ICIs. In order to answer these questions, immunological analyses using clinical specimens are essential. It is therefore important to develop effective therapeutic strategies combining ICIs with other anti-cancer drugs and to identify biomarkers that can differentiate responders from non- responders by clarifying the immune suppressive networks in the TME.
List of papers published in 2022
Journal
1. Hiramatsu H, Nosaka K, Kusumoto S, Nakano N, Choi I, Yoshimitsu M, Imaizumi Y, Hidaka M, Sasaki H, Makiyama J, Ohtsuka E, Jo T, Ogata M, Ito A, Yonekura K, Tatetsu H, Kato T, Kawakita T, Suehiro Y, Ishitsuka K, Iida S, Matsutani T, Nishikawa H, Utsunomiya A, Ueda R, Ishida T. Landscape of immunoglobulin heavy chain γ gene class switch recombination in patients with adult T-cell leukemia-lymphoma. Haematologica, 108:1173-1178, 2023
2. Sugiyama D, Hinohara K, Nishikawa H. Significance of regulatory T cells in cancer immunology and immunotherapy. Experimental dermatology, 32:256-263, 2023
3. Kato S, Maeda Y, Sugiyama D, Watanabe K, Nishikawa H, Hinohara K. The cancer epigenome: Non-cell autonomous player in tumor immunity. Cancer science, 114:730-740, 2023
4. Itahashi K, Irie T, Yuda J, Kumagai S, Tanegashima T, Lin YT, Watanabe S, Goto Y, Suzuki J, Aokage K, Tsuboi M, Minami Y, Ishii G, Ohe Y, Ise W, Kurosaki T, Suzuki Y, Koyama S, Nishikawa H. BATF epigenetically and transcriptionally controls the activation program of regulatory T cells in human tumors. Science immunology, 7:eabk0957, 2022
5. Yukami H, Kawazoe A, Lin YT, Koyama S, Fukuoka S, Hara H, Takahashi N, Kojima T, Asayama M, Yoshii T, Bando H, Kotani D, Nakamura Y, Kuboki Y, Mishima S, Wakabayashi M, Kuwata T, Goto M, Higuchi K, Yoshino T, Doi T, Nishikawa H, Shitara K. Updated Efficacy Outcomes of Anti-PD-1 Antibodies plus Multikinase Inhibitors for Patients with Advanced Gastric Cancer with or without Liver Metastases in Clinical Trials. Clinical cancer research, 28:3480-3488, 2022
6. Tanaka N, Mori S, Kiyotani K, Ota Y, Gotoh O, Kusumoto S, Nakano N, Suehiro Y, Ito A, Choi I, Ohtsuka E, Hidaka M, Nosaka K, Yoshimitsu M, Imaizumi Y, Iida S, Utsunomiya A, Noda T, Nishikawa H, Ueda R, Ishida T. Genomic determinants impacting the clinical outcome of mogamulizumab treatment for adult T-cell leukemia/lymphoma. Haematologica, 107:2418-2431, 2022
7. Goda N, Sasada S, Shigematsu H, Masumoto N, Arihiro K, Nishikawa H, Sakaguchi S, Okada M, Kadoya T. The ratio of CD8 + lymphocytes to tumor-infiltrating suppressive FOXP3 + effector regulatory T cells is associated with treatment response in invasive breast cancer. Discover Oncology, 13:27, 2022
8. Kobayashi T, Kumagai S, Doi R, Afonina E, Koyama S, Nishikawa H. Isolation of tumor-infiltrating lymphocytes from preserved human tumor tissue specimens for downstream characterization. STAR protocols, 3:101557, 2022
9. Itahashi K, Irie T, Nishikawa H. Regulatory T-cell development in the tumor microenvironment. European journal of immunology, 52:1216-1227, 2022
10. Kelkka T, Tyster M, Lundgren S, Feng X, Kerr C, Hosokawa K, Huuhtanen J, Keränen M, Patel B, Kawakami T, Maeda Y, Nieminen O, Kasanen T, Aronen P, Yadav B, Rajala H, Nakazawa H, Jaatinen T, Hellström-Lindberg E, Ogawa S, Ishida F, Nishikawa H, Nakao S, Maciejewski J, Young NS, Mustjoki S. Anti-COX-2 autoantibody is a novel biomarker of immune aplastic anemia. Leukemia, 36:2317-2327, 2022
11. Nagaharu K, Kojima Y, Hirose H, Minoura K, Hinohara K, Minami H, Kageyama Y, Sugimoto Y, Masuya M, Nii S, Seki M, Suzuki Y, Tawara I, Shimamura T, Katayama N, Nishikawa H, Ohishi K. A bifurcation concept for B-lymphoid/plasmacytoid dendritic cells with largely fluctuating transcriptome dynamics. Cell reports, 40:111260, 2022
12. Aoki C, Imai K, Mizutani T, Sugiyama D, Miki R, Koya Y, Kobayashi T, Ushida T, Iitani Y, Nakamura N, Owaki T, Nishikawa H, Toyokuni S, Kajiyama H, Kotani T. Molecular hydrogen has a positive impact on pregnancy maintenance through enhancement of mitochondrial function and immunomodulatory effects on T cells. Life sciences, 308:120955, 2022
13. Terasaki F, Ohgi K, Sugiura T, Okamura Y, Ito T, Yamamoto Y, Ashida R, Yamada M, Otsuka S, Aramaki T, Uesaka K. Portal vein thrombosis after right hepatectomy: impact of portal vein resection and morphological changes of the portal vein. HPB, 24:1129-1137, 2022
14. Iwasawa T, Kosaka T, Morita S, Mikami S, Nakamura K, Hongo H, Nishihara H, Oya M. A Japanese case of castration-resistant prostate cancer with BRCA2 and RB1 co-loss and TP53 mutation: a case report. BMC medical genomics, 15:138, 2022
15. Iwasawa T, Kosaka T, Yasumizu Y, Hongo H, Yanai Y, Baba Y, Matsumoto K, Nakamura K, Nishihara H, Oya M. Characterizing cyclin-dependent kinase 12(CDK12)-altered aggressive prostate cancer: a twelve-case series. International journal of clinical oncology, 27:1867-1873, 2022
