Annual Report 2022
Department of Genetic Medicine and Services
Teruhiko Yoshida, Makoto Hirata, Hourin Cho, Noriko Tanabe, Tomoko Watanabe, Sayuri Hiraoka, Shigenobu Suzuki, Mitsuya Ishikawa, Nobuyoshi Hiraoka, Shigeki Sekine, Taisuke Mori, Kuniko Sunami, Noboru Yamamoto, Nozomu Kobayashi, Satoyo Oda, Takashi Kohno, Masahiro Goto, Hitoshi Ichikawa, Hiromi Sakamoto, Mamoru Kato, Eisaku Furukawa.
Introduction
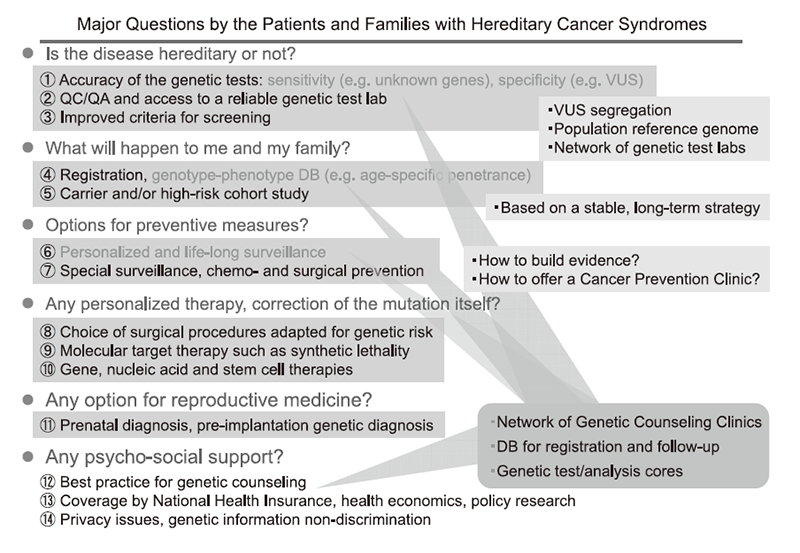
It has been estimated that roughly 5% of all cancer cases are caused by a highly penetrant monogenic germline variant. The major causative genes for most hereditary cancer syndromes were identified in the 1990s. Since then, genetic diagnosis has been considered as a part of standard medical care in oncology clinics. The National Cancer Center Hospital (NCCH) launched the Outpatient Genetic Counseling Clinic in 1998 as part of a collaboration with the NCC Research Institute, specifically the Fundamental Innovative Oncology Core (FIOC). However, several issues remain to be addressed in cancer medical genetics, as summarized in Figure 1.
The Team and What We Do
The Department of Genetic Medicine and Services (GeMS) consists of 11 doctors, seven researchers, two genetic counselors, and one clinical laboratory technician. All of these staff hold a concurrent post of another department and research division, except the genetic counselors.
As shown on the NCCH website, the aim and mission of the clinical service of the Outpatient Genetic Counseling Clinic, which are the major routine clinical activities of our department, are:
1) To provide consultation and appropriate medical and genetic information (that is, genetic counseling) to anyone who has a concern related to heredity cancer.
2) To provide genetic testing when appropriate.
3) To support early diagnosis and treatment based on family history and/or genetic test results.
4) To discuss clinical sequencing results of patients by participating in the Expert panel.
5) To provide genetic counseling and germline confirmatory testing upon request from each clinical department regarding germline-related findings obtained through cancer comprehensive genomic panel testing.
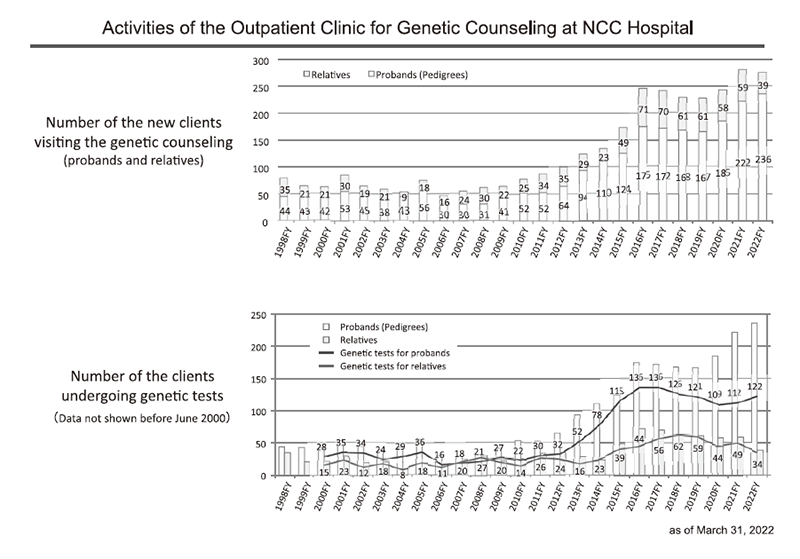
For the period of April 2022 to March 2023, 433 clients and their relatives visited the clinic. Of these, 236 families and 275 clients newly visited this period. In total, 2,317 families have visited the clinic since its inception in 1998. Details are shown in Table 1 and Figure 2.
Table 1. Number of new clients (Apr. 2022 – Mar. 2023)

Research Activities
Although at least one causative gene has been identified for each of the major hereditary cancer syndromes, overall sensitivity of the current genetic tests is far from 100% and may be around 70-80%, even for the cases that meet clinical and/or screening criteria for hereditary cancer syndromes. The false negative cases may include both inadequate technical sensitivity of the current genetic test methods on the established causative genes (allelic heterogeneity) and also yet-to-be-identified genes representing locus heterogeneity. There has been great expectation that the introduction of next generation sequencers (NGS) would change the situation. The staff of the Department of GeMS have established a new Common Protocol to perform NGS-based germline clinical sequencing for patients with negative test results by conventional genetic tests, who would represent a part of the Undiagnosed Disease Patients in the oncology field. The Common Protocol has been adopted by other hospitals and institutions in a long-standing multi-institute collaborative research group based on the National Cancer Center Research and Development Fund and its predecessor. In addition to whole exome/genome sequencing (WES/WGS), a multi-gene panel has been developed based on Agilent SureSelect technology.
Clinical Trials
We have participated in an expert panel of clinical sequencing conducted by health insurance systems since May 29, 2019. The staffs of the Department of GeMS have not acted as principal investigators of clinical intervention trials but have participated in prospective clinical studies to develop and evaluate the CGP tests for the TOP2 CGP panel conducted by the Japanese Children's Cancer Study Group (JCCG) since 2021. Furthermore, we are actively engaged in the cancer genome analysis research project within the framework of the AMED Innovative Cancer Control program, aligning with the Action Plan for Whole Genome Analysis 2022. Our efforts encompass a range of genetic-related research tasks, spanning from the acquisition of informed consent to patient-centric initiatives.
Education
The Department has accepted attendees for outpatient genetic counseling, so that they could be eligible to take the examination for clinical geneticists and certified genetic counselors acknowledged by the Japanese Society of Human Genetics and the Japanese Society of Genetic Counseling. In 2022, five doctors registered as trainees for clinical geneticists in the education committee of clinical geneticists. We accepted one resident and one oncologist for training. Furthermore, we accepted six students of the genetic counseling course in the Graduate School of Health and Welfare Sciences, International University of Health and Welfare.
Future Prospects
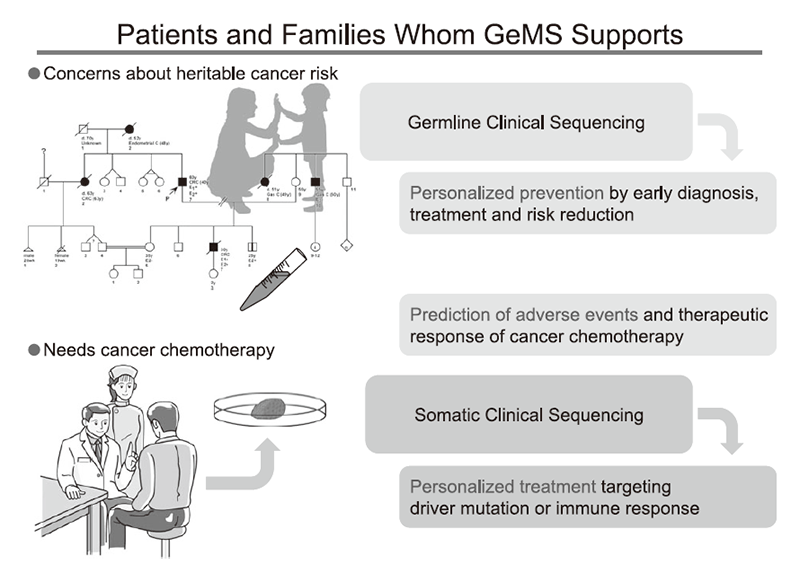
The Department of GeMS was launched in November 2015. Although this section reports on the routine clinical activity of the Department and clinical research associated directly with the Outpatient Genetic Counseling Clinic, the scope and mission of the Department extend beyond hereditary cancer syndromes and include support of the genomic biomarker-driven personalized cancer treatments offered by other clinical departments (Figure 3). The core technology of the new discipline, also known as a precision medicine, is next-generation sequencers, which would bring massive amounts of genomic data to cancer diagnosis, treatment and prevention. The crux of this emerging opportunity is how to convert the sequence data to clinically valid and useful knowledge, which could include incidental or secondary findings. The Department of GeMS is expected to support other departments in the era of genomic medicine.
List of papers published in 2022
Journal
1. Naka T, Hashimoto T, Cho H, Tanabe N, Yoshida T, Yatabe Y, Yoshikawa T, Abe S, Sekine S. Sporadic and Familial Adenomatous Polyposis-associated Foveolar-type Adenoma of the Stomach. The American journal of surgical pathology, 47:91-101, 2023
2. Fukushi G, Yamada M, Kakugawa Y, Gotoh M, Tanabe N, Ushiama M, Watanabe T, Yamazaki T, Matsumoto M, Hirata M, Nakajima T, Sugano K, Yoshida T, Matsuda T, Igarashi Y, Saito Y. Genotype-phenotype correlation of small-intestinal polyps on small-bowel capsule endoscopy in familial adenomatous polyposis. Gastrointestinal endoscopy, 97:59-68.e7, 2023
3. Murano T, Ikematsu H, Shinmura K, Okumura K, Kuwata T, Ushiama M, Yoshida T, Takashima K, Nakajo K, Kadota T, Yoda Y, Oono Y, Yano T. Endoscopic management of familial adenomatous polyposis targeting colorectal lesions greater than 5 mm in size: a single-center retrospective study. Familial cancer, 22:83-89, 2023
4. Naka T, Hashimoto T, Yoshida T, Yatabe Y, Sekine S. KLF4 c.A1322C Mutation Is a Consistent and Specific Genetic Feature of Raspberry-like Foveolar-type Adenoma of the Stomach. The American journal of surgical pathology, 47:521-523, 2023
5. Akiyama M, Sakaue S, Takahashi A, Ishigaki K, Hirata M, Matsuda K, Momozawa Y, Okada Y, Ninomiya T, Terao C, Murakami Y, Kubo M, Kamatani Y. Genome-wide association study reveals BET1L associated with survival time in the 137,693 Japanese individuals. Communications biology, 6:143, 2023
6. Usui Y, Taniyama Y, Endo M, Koyanagi YN, Kasugai Y, Oze I, Ito H, Imoto I, Tanaka T, Tajika M, Niwa Y, Iwasaki Y, Aoi T, Hakozaki N, Takata S, Suzuki K, Terao C, Hatakeyama M, Hirata M, Sugano K, Yoshida T, Kamatani Y, Nakagawa H, Matsuda K, Murakami Y, Spurdle AB, Matsuo K, Momozawa Y. Helicobacter pylori, Homologous-Recombination Genes, and Gastric Cancer. The New England journal of medicine, 388:1181-1190, 2023
7. Omura T, Takahashi M, Ohno M, Miyakita Y, Yanagisawa S, Tamura Y, Kikuchi M, Kawauchi D, Nakano T, Hosoya T, Igaki H, Satomi K, Yoshida A, Sunami K, Hirata M, Shimoi T, Sudo K, Okuma HS, Yonemori K, Suzuki H, Ichimura K, Narita Y. Clinical Application of Comprehensive Genomic Profiling Tests for Diffuse Gliomas. Cancers, 14:2454, 2022
8. Ida H, Koyama T, Mizuno T, Sunami K, Kubo T, Sudo K, Tao K, Hirata M, Yonemori K, Kato K, Okusaka T, Ohe Y, Matsui Y, Yamazaki N, Ogawa C, Kawai A, Narita Y, Esaki M, Yamamoto N. Clinical utility of comprehensive genomic profiling tests for advanced or metastatic solid tumor in clinical practice. Cancer science, 113:4300-4310, 2022
9. Udagawa C, Nakano MH, Yoshida T, Ohe Y, Kato K, Mushiroda T, Zembutsu H. Association between genetic variants and the risk of nivolumab-induced immune-related adverse events. Pharmacogenomics, 23:887-901, 2022
10. Kohno T, Kato M, Kohsaka S, Sudo T, Tamai I, Shiraishi Y, Okuma Y, Ogasawara D, Suzuki T, Yoshida T, Mano H. C-CAT: The National Datacenter for Cancer Genomic Medicine in Japan. Cancer discovery, 12:2509-2515, 2022
11. Udagawa C, Kuah S, Shimoi T, Kato K, Yoshida T, Nakano MH, Shimo A, Kojima Y, Yoshie R, Tsugawa K, Mushiroda T, Tan EY, Zembutsu H. Replication Study for the Association of Five SNPs Identified by GWAS and Trastuzumab-Induced Cardiotoxicity in Japanese and Singaporean Cohorts. Biological & pharmaceutical bulletin, 45:1198-1202, 2022
12. Fukuhara S, Oshikawa-Kumade Y, Kogure Y, Shingaki S, Kariyazono H, Kikukawa Y, Koya J, Saito Y, Tabata M, Yoshifuji K, Mizuno K, Miyagi-Maeshima A, Matsushita H, Sugiyama M, Ogawa C, Inamoto Y, Fukuda T, Sugano M, Yamauchi N, Minami Y, Hirata M, Yoshida T, Kohno T, Kohsaka S, Mano H, Shiraishi Y, Ogawa S, Izutsu K, Kataoka K. Feasibility and clinical utility of comprehensive genomic profiling of hematological malignancies. Cancer science, 113:2763-2777, 2022
13. Imai M, Nakamura Y, Sunami K, Kage H, Komine K, Koyama T, Amano T, Ennishi D, Kanai M, Kenmotsu H, Maeda T, Morita S, Sakai D, Bando H, Makiyama A, Suzuki T, Hirata M, Kohsaka S, Tsuchihara K, Naito Y, Yoshino T. Expert panel consensus recommendations on the use of circulating tumor DNA assays for patients with advanced solid tumors. Cancer science, 113:3646-3656, 2022
14. Hamamoto R, Koyama T, Kouno N, Yasuda T, Yui S, Sudo K, Hirata M, Sunami K, Kubo T, Takasawa K, Takahashi S, Machino H, Kobayashi K, Asada K, Komatsu M, Kaneko S, Yatabe Y, Yamamoto N. Introducing AI to the molecular tumor board: one direction toward the establishment of precision medicine using large-scale cancer clinical and biological information. Experimental hematology & oncology, 11:82, 2022
15. Naito Y, Sunami K, Kage H, Komine K, Amano T, Imai M, Koyama T, Ennishi D, Kanai M, Kenmotsu H, Maeda T, Morita S, Sakai D, Watanabe K, Shirota H, Kinoshita I, Yoshioka M, Mamesaya N, Ito M, Kohsaka S, Saigusa Y, Yamamoto K, Hirata M, Tsuchihara K, Yoshino T. Concordance Between Recommendations From Multidisciplinary Molecular Tumor Boards and Central Consensus for Cancer Treatment in Japan. JAMA network open, 5:e2245081, 2022
16. Saito A, Tanioka M, Hirata M, Watanabe T, Odaka Y, Shimoi T, Sudo K, Noguchi E, Ishikawa M, Yonemori K. Case report: Response to platinum agents and poly (adenosine diphosphate-ribose) polymerase inhibitor in a patient with BRCA1 c.5096G>A (R1699Q) intermediate-risk variant. Cancer treatment and research communications, 32:100587, 2022
17. Takamiya A, Ishibashi Y, Makise N, Hirata M, Ushiku T, Tanaka S, Kobayashi H. Imaging characteristics of NTRK-rearranged spindle cell neoplasm of the soft tissue: A case report. Journal of orthopaedic science, S0949-2658(21)00367-5, 2022
18. Yagi Y, Abeto N, Shiraishi J, Miyata C, Inoue S, Murakami H, Nakashima M, Sugano K, Ushiama M, Yoshida T, Yamazawa K. A novel pathogenic variant of the FH gene in a family with hereditary leiomyomatosis and renal cell carcinoma. Human genome variation, 9:3, 2022
19. Tomida A, Chiyonobu T, Tokuda S, Miyachi M, Murashima K, Hirata M, Nakagawa M, Iehara T, Kuroda J, Takayama K. Pleomorphic rhabdomyosarcoma in a young adult harboring a novel germline MSH2 variant. Human genome variation, 9:8, 2022
20. Momozawa Y, Sasai R, Usui Y, Shiraishi K, Iwasaki Y, Taniyama Y, Parsons MT, Mizukami K, Sekine Y, Hirata M, Kamatani Y, Endo M, Inai C, Takata S, Ito H, Kohno T, Matsuda K, Nakamura S, Sugano K, Yoshida T, Nakagawa H, Matsuo K, Murakami Y, Spurdle AB, Kubo M. Expansion of Cancer Risk Profile for BRCA1 and BRCA2 Pathogenic Variants. JAMA oncology, 8:871-878, 2022
21. Sekine Y, Iwasaki Y, Aoi T, Endo M, Hirata M, Kamatani Y, Matsuda K, Sugano K, Yoshida T, Murakami Y, Fukui T, Akamatsu S, Ogawa O, Nakagawa H, Numakura K, Narita S, Habuchi T, Momozawa Y. Different risk genes contribute to clear cell and non-clear cell renal cell carcinoma in 1532 Japanese patients and 5996 controls. Human molecular genetics, 31:1962-1969, 2022
22. Usui Y, Iwasaki Y, Matsuo K, Endo M, Kamatani Y, Hirata M, Sugano K, Yoshida T, Matsuda K, Murakami Y, Maeda Y, Nakagawa H, Momozawa Y. Association between germline pathogenic variants in cancer-predisposing genes and lymphoma risk. Cancer science, 113:3972-3979, 2022
23. Komatsu M, Ichikawa H, Chiwaki F, Sakamoto H, Komatsuzaki R, Asaumi M, Tsunoyama K, Fukagawa T, Matsushita H, Boku N, Matsusaki K, Takeshita F, Yoshida T, Sasaki H. ARHGAP-RhoA signaling provokes homotypic adhesion-triggered cell death of metastasized diffuse-type gastric cancer. Oncogene, 41:4779-4794, 2022
24. Mishra A, Malik R, Hachiya T, Jürgenson T, Namba S, Posner DC, Kamanu FK, Koido M, Le Grand Q, Shi M, He Y, Georgakis MK, Caro I, Krebs K, Liaw YC, Vaura FC, Lin K, Winsvold BS, Srinivasasainagendra V, Parodi L, Bae HJ, Chauhan G, Chong MR, Tomppo L, Akinyemi R, Roshchupkin GV, Habib N, Jee YH, Thomassen JQ, Abedi V, Cárcel-Márquez J, Nygaard M, Leonard HL, Yang C, Yonova-Doing E, Knol MJ, Lewis AJ, Judy RL, Ago T, Amouyel P, Armstrong ND, Bakker MK, Bartz TM, Bennett DA, Bis JC, Bordes C, Børte S, Cain A, Ridker PM, Cho K, Chen Z, Cruchaga C, Cole JW, de Jager PL, de Cid R, Endres M, Ferreira LE, Geerlings MI, Gasca NC, Gudnason V, Hata J, He J, Heath AK, Ho YL, Havulinna AS, Hopewell JC, Hyacinth HI, Inouye M, Jacob MA, Jeon CE, Jern C, Kamouchi M, Keene KL, Kitazono T, Kittner SJ, Konuma T, Kumar A, Lacaze P, Launer LJ, Lee KJ, Lepik K, Li J, Li L, Manichaikul A, Markus HS, Marston NA, Meitinger T, Mitchell BD, Montellano FA, Morisaki T, Mosley TH, Nalls MA, Nordestgaard BG, O’Donnell MJ, Okada Y, Onland-Moret NC, Ovbiagele B, Peters A, Psaty BM, Rich SS, Rosand J, Sabatine MS, Sacco RL, Saleheen D, Sandset EC, Salomaa V, Sargurupremraj M, Sasaki M, Satizabal CL, Schmidt CO, Shimizu A, Smith NL, Sloane KL, Sutoh Y, Sun YV, Tanno K, Tiedt S, Tatlisumak T, Torres-Aguila NP, Tiwari HK, Trégouët DA, Trompet S, Tuladhar AM, Tybjærg-Hansen A, van Vugt M, Vibo R, Verma SS, Wiggins KL, Wennberg P, Woo D, Wilson PWF, Xu H, Yang Q, Yoon K, Millwood IY, Gieger C, Ninomiya T, Grabe HJ, Jukema JW, Rissanen IL, Strbian D, Kim YJ, Chen PH, Mayerhofer E, Howson JMM, Irvin MR, Adams H, Wassertheil-Smoller S, Christensen K, Ikram MA, Rundek T, Worrall BB, Lathrop GM, Riaz M, Simonsick EM, Kõrv J, França PHC, Zand R, Prasad K, Frikke-Schmidt R, de Leeuw FE, Liman T, Haeusler KG, Ruigrok YM, Heuschmann PU, Longstreth WT, Jung KJ, Bastarache L, Paré G, Damrauer SM, Chasman DI, Rotter JI, Anderson CD, Zwart JA, Niiranen TJ, Fornage M, Liaw YP, Seshadri S, Fernández-Cadenas I, Walters RG, Ruff CT, Owolabi MO, Huffman JE, Milani L, K. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature, 611:115-123, 2022
25. Tsuda N, Tian Y, Fujimoto M, Kuramoto J, Makiuchi S, Ojima H, Gotoh M, Hiraoka N, Yoshida T, Kanai Y, Arai E. DNA methylation status of the SPHK1 and LTB genes underlies the clinicopathological diversity of non-alcoholic steatohepatitis-related hepatocellular carcinomas. Journal of cancer research and clinical oncology, 2022
26. Liu X, Yamaguchi K, Takane K, Zhu C, Hirata M, Hikiba Y, Maeda S, Furukawa Y, Ikenoue T. Cancer-associated IDH mutations induce Glut1 expression and glucose metabolic disorders through a PI3K/Akt/mTORC1-Hif1α axis. PloS one, 16:e0257090, 2021
