Annual Report 2022
Division of Medical Ai Research and Development
Ryuji Hamamoto, Syuzo Kaneko, Kazuma Kobayashi, Masaaki Komatsu, Ken Asada, Satoshi Takahashi, Ken Takasawa, Masayoshi Yamada, Masamichi Takahashi, Kyoko Fujioka, Noriko Ikawa, Hiroko Kondo, Shigemi Yamada, Hidenori Machino, Amina Bolatkan, Norio Shinkai, Akira Sakai, Rina Aoyama, Nobuji Kouno
Introduction
Our division launched a pioneering large-scale project for medical artificial intelligence (AI) in Japan called "Development of an Integrated Cancer Medical System Using AI” after the decision to promote research and development of AI as a national policy based on the "Fifth Science and Technology Basic Plan" approved by the Cabinet in January 2016. Currently, as the core of medical AI research and development at the Tsukiji Campus, and as a center for medical AI research and development in Japan, the project has been actively promoted. In particular, research activities are focused on the following two points: 1) to promote research for the benefit of patients without falling into research for research's sake, aiming at clinical applications, and 2) to construct an integrated database that contains a wealth of high-quality medical information.
Research Activities
1. Development of new cancer diagnostic methods using AI technology
In close collaboration with physicians from various departments and divisions of the National Cancer Center Hospital, we conducted research and development of AI technology to support endoscopic diagnosis, radiological imaging, ultrasound diagnosis, and skin imaging diagnosis. In particular, the endoscopic diagnosis support AI that we developed has already been clinically applied since its approval by the pharmaceutical affairs bodies in 2020 and conformed to the requirements of the CE Mark. We have further succeeded in developing an AI that automatically and robustly predicts pathological diagnosis based on the revised Vienna Classification. This achievement shows that the developed AI system can assist endoscopists in real time, avoid false positives during colonoscopy, and improve differential diagnosis of colorectal cancer. This was published as a scientific paper (J Gastroenterol. 2022 Nov;57(11):879-889). We have continuously met with the PMDA to obtain regulatory approval as a medical device.
2. Development of the Medical AI Development Platform
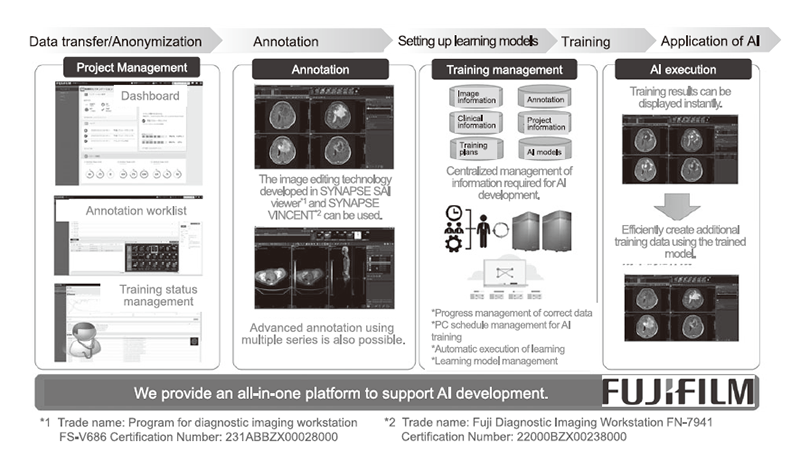
We developed an AI development support platform, a research infrastructure system that enables physicians to develop AI technologies, in collaboration with Fujifilm, and commercialized it under the name "Synapse Creative Space" on April 5, 2022 (Figure 1). With the functions of this platform, constructing both software and hardware environments for AI development and acquiring advanced engineering knowledge required for designing learning models are no longer necessary, which have been required for research and development of AI technologies to support diagnostic imaging, thereby reducing the processing and management of a large number of training data. The system also saves physicians from spending a huge amount of time on processing and managing a large amount of training data.
3. Construction of the world's largest integrated database of lung cancer data oriented toward AI analysis and development of a multi-omics analysis platform using AI technology
As part of the PRISM project, we have constructed one of the world's largest integrated lung cancer databases oriented toward AI analysis, with more than 1,700 cases. In conjunction with the construction of the integrated database, we have also built a system that efficiently collects medical information from electronic medical records and pathology systems, as well as medical image information from various modalities. In addition, three AI technology-based platforms have been developed to efficiently and accurately analyze large-scale omics data related to cancer: "Multiomics_Analyzer," "Reverse Phase Protein Array (RPPA) Analysis," and "Modified_Diet_Networks." These platforms were gadgetized under their respective names and installed in the open platform "Mine".
Education
A total of four graduate students from Kyoto University, Tokyo Medical and Dental University, and Showa University belonged to this division and provided research guidance. Members of the division also actively participated in young researchers' seminars organized by the institute.
Future Prospects
1. We will continue to promote research and development of novel cancer diagnosis support AI in collaboration with various departments and divisions of the NCCH, aiming for clinical application.
2. We will promote research utilization of the AI development platform in multiple research themes within the National Cancer Center Japan, aiming for regulatory approval of medical devices.
3. We will continue to develop an omics analysis platform using AI technology while expanding the integrated database for lung cancer and working on the construction of integrated databases for cancer types other than lung cancer.
List of papers published in 2022
Journal
1. Asami Y, Kobayashi Kato M, Hiranuma K, Matsuda M, Shimada Y, Ishikawa M, Koyama T, Komatsu M, Hamamoto R, Nagashima M, Terao Y, Itakura A, Kohno T, Sekizawa A, Matsumoto K, Kato T, Shiraishi K, Yoshida H. Utility of molecular subtypes and genetic alterations for evaluating clinical outcomes in 1029 patients with endometrial cancer. British journal of cancer, 128:1582-1591, 2023
2. Hossain E, Abdelrahim M, Tanasescu A, Yamada M, Kondo H, Yamada S, Hamamoto R, Marugame A, Saito Y, Bhandari P. Performance of a novel computer-aided diagnosis system in the characterization of colorectal polyps, and its role in meeting Preservation and Incorporation of Valuable Endoscopic Innovations standards set by the American Society of Gastrointestinal Endoscopy. DEN open, 3:e178, 2023
3. Ito T, Takayanagi D, Sekine S, Hashimoto T, Shimada Y, Matsuda M, Yamada M, Hamamoto R, Kato T, Shida D, Kanemitsu Y, Boku N, Kohno T, Takashima A, Shiraishi K. Comparison of clinicopathological and genomic profiles in anal squamous cell carcinoma between Japanese and Caucasian cohorts. Scientific reports, 13:3587, 2023
4. Hamamoto R, Takasawa K, Shinkai N, Machino H, Kouno N, Asada K, Komatsu M, Kaneko S. Analysis of super-enhancer using machine learning and its application to medical biology. Briefings in bioinformatics, 24:bbad107, 2023
5. Shirasawa M, Yoshida T, Shiraishi K, Takigami A, Takayanagi D, Imabayashi T, Matsumoto Y, Masuda K, Shinno Y, Okuma Y, Goto Y, Horinouchi H, Yotsukura M, Yoshida Y, Nakagawa K, Tsuchida T, Hamamoto R, Yamamoto N, Motoi N, Kohno T, Watanabe SI, Ohe Y. Identification of inflamed-phenotype of small cell lung cancer leading to the efficacy of anti-PD-L1 antibody and chemotherapy. Lung cancer (Amsterdam, Netherlands), 179:107183, 2023
6. Kim K, Ryu TY, Jung E, Han TS, Lee J, Kim SK, Roh YN, Lee MS, Jung CR, Lim JH, Hamamoto R, Lee HW, Hur K, Son MY, Kim DS, Cho HS. Epigenetic regulation of SMAD3 by histone methyltransferase SMYD2 promotes lung cancer metastasis. Experimental & molecular medicine, 55:952-964, 2023
7. Yamada M, Shino R, Kondo H, Yamada S, Takamaru H, Sakamoto T, Bhandari P, Imaoka H, Kuchiba A, Shibata T, Saito Y, Hamamoto R. Robust automated prediction of the revised Vienna Classification in colonoscopy using deep learning: development and initial external validation. Journal of gastroenterology, 57:879-889, 2022
8. Hashimoto T, Takayanagi D, Yonemaru J, Naka T, Nagashima K, Yatabe Y, Shida D, Hamamoto R, Kleeman SO, Leedham SJ, Maughan T, Takashima A, Shiraishi K, Sekine S. Clinicopathological and molecular characteristics of RSPO fusion-positive colorectal cancer. British journal of cancer, 127:1043-1050, 2022
9. Hamamoto R, Koyama T, Kouno N, Yasuda T, Yui S, Sudo K, Hirata M, Sunami K, Kubo T, Takasawa K, Takahashi S, Machino H, Kobayashi K, Asada K, Komatsu M, Kaneko S, Yatabe Y, Yamamoto N. Introducing AI to the molecular tumor board: one direction toward the establishment of precision medicine using large-scale cancer clinical and biological information. Experimental hematology & oncology, 11:82, 2022
10. Dozen A, Shozu K, Shinkai N, Ikawa N, Aoyama R, Machino H, Asada K, Yoshida H, Kato T, Hamamoto R, Kaneko S, Komatsu M. Tumor Suppressive Role of the PRELP Gene in Ovarian Clear Cell Carcinoma. Journal of personalized medicine, 12:1999, 2022
11. Ono S, Komatsu M, Sakai A, Arima H, Ochida M, Aoyama R, Yasutomi S, Asada K, Kaneko S, Sasano T, Hamamoto R. Automated Endocardial Border Detection and Left Ventricular Functional Assessment in Echocardiography Using Deep Learning. Biomedicines, 10:1082, 2022
12. Hamamoto R, Takasawa K, Machino H, Kobayashi K, Takahashi S, Bolatkan A, Shinkai N, Sakai A, Aoyama R, Yamada M, Asada K, Komatsu M, Okamoto K, Kameoka H, Kaneko S. Application of non-negative matrix factorization in oncology: one approach for establishing precision medicine. Briefings in bioinformatics, 23:bbac246, 2022
13. Hopkins J, Asada K, Leung A, Papadaki V, Davaapil H, Morrison M, Orita T, Sekido R, Kosuge H, Reddy MA, Kimura K, Mitani A, Tsumoto K, Hamamoto R, Sagoo MS, Ohnuma SI. PRELP Regulates Cell-Cell Adhesion and EMT and Inhibits Retinoblastoma Progression. Cancers, 14:4926, 2022
14. Kukita A, Sone K, Kaneko S, Kawakami E, Oki S, Kojima M, Wada M, Toyohara Y, Takahashi Y, Inoue F, Tanimoto S, Taguchi A, Fukuda T, Miyamoto Y, Tanikawa M, Mori-Uchino M, Tsuruga T, Iriyama T, Matsumoto Y, Nagasaka K, Wada-Hiraike O, Oda K, Hamamoto R, Osuga Y. The Histone Methyltransferase SETD8 Regulates the Expression of Tumor Suppressor Genes via H4K20 Methylation and the p53 Signaling Pathway in Endometrial Cancer Cells. Cancers, 14:5367, 2022
15. Shozu K, Kaneko S, Shinkai N, Dozen A, Kosuge H, Nakakido M, Machino H, Takasawa K, Asada K, Komatsu M, Tsumoto K, Ohnuma SI, Hamamoto R. Repression of the PRELP gene is relieved by histone deacetylase inhibitors through acetylation of histone H2B lysine 5 in bladder cancer. Clinical epigenetics, 14:147, 2022
