Home > Information > Investigator-initiated clinical studies to be launched following confirmation of tumor shrinkage induced by the targeted protein degrader E7820 using J-PDX (Japanese cancer patient-derived tissue transplantation models)
Investigator-initiated clinical studies to be launched following confirmation of tumor shrinkage induced by the targeted protein degrader E7820 using J-PDX (Japanese cancer patient-derived tissue transplantation models)Aiming to establish a drug discovery and development system that accelerates the development of new anticancer drugs
September 12, 2024
National Cancer Center
Eisai Co., Ltd.
Highlights
- Tumor-agnostic efficacy evaluation of Eisai's targeted protein degrader E7820 was performed using patient-derived xenograft (PDX) models created by transplanting tumor tissue derived from patients into immunodeficient mice (pancreatic cancer, bile duct cancer, gastric cancer, and uterine cancer), which observed tumor shrinkage in 38.1% for overall, 58.3% for bile duct cancer, and 55.6% for uterine cancer.
- Since PDX models have high predictive accuracy for clinical outcomes and high similarity in responsiveness with clinical data, this result indicates that E7820 is effective in shrinking tumors of the bile duct and uterus.
- Whole-exome sequencing in the PDX models identified genetic mutations that may be used as predictive biomarkers of response to E7820.
- The findings were published in the peer-reviewed journal npj Precision Oncology.
- Based on the cancer types and predictive biomarkers associated with E7820 efficacy revealed by this study, an investigator-initiated clinical study will be launched at the National Cancer Center Hospital and the National Cancer Center Hospital East for the purposes of safety evaluation including the tolerability in Japanese patients, and exploratory efficacy evaluation.
- The use of the National Cancer Center’s library of J-PDX (Japanese cancer patient-derived tissue transplantation models) for drug discovery and development has made it possible to evaluate the drug efficacy in each cancer type, identify predictive biomarkers, and launch a clinical study in one seamless process. Through further utilization of this process, both parties will aim to establish a drug discovery and development system that accelerates the development of new anticancer drugs.
Summary
The National Cancer Center (Location: Tokyo; President: Hitoshi Nakagama) and Eisai Co., Ltd. (Headquarters: Tokyo; CEO: Haruo Naito; "Eisai") have been carrying out since 2021 the “Basic research on the drug discovery and development to accelerate development of anticancer drugs in treatment of patients with rare cancers and refractory cancers” aimed at establishing a drug discovery and development system that accelerates the development of new anticancer drugs with funding under the program “Cyclic Innovation for Clinical Empowerment (CiCLE)” led by the Japan Agency for Medical Research and Development (AMED).
In this research, the National Cancer Center and Eisai used the library of J-PDX,1) animal models in which tumor tissue derived from Japanese cancer patients was transplanted into immunodeficient mice, established by the National Cancer Center Research Institute (Director: Hiroyuki Mano) to conduct a non-clinical study to evaluate the efficacy of Eisai’s new drug candidate E7820 in 42 PDX models (12 pancreatic cancer models, 12 bile duct cancer models, 9 gastric cancer models, and 9 uterine cancer models).
The results showed that the administration of E7820 at 100 mg/kg led to tumor shrinkage in 16 of the 42 models (38.1%), including 7 of the 12 bile duct cancer models (58.3%) and 5 of the 9 uterine cancer models (55.6%). Since PDX models have high predictive accuracy for clinical outcomes and high similarity in responsiveness with clinical data, the results of this study indicate that E7820 is effective in shrinking tumors of the bile duct and uterus.
Furthermore, to explore the biomarkers2) correlated with the tumor-shrinking effect of E7820, whole-exome sequencing3) was performed in the PDX models, which observed frequent mutations in genes related to homologous recombination repair (HRR)4) (one of the DNA repair mechanisms) such as BRCA1, BRCA2, and ATM in the PDX models that showed tumor shrinkage, suggesting that mutations in these genes may be used as biomarkers of response to E7820.
The findings were published in the peer-reviewed journal npj Precision Oncology.
Based on these results, the National Cancer Center Hospital (Director: Yasuyuki Seto) and the Hospital East (Director: Toshihiko Doi) have launched a phase I investigator-initiated clinical study5) (NCCH2303) to evaluate the safety and efficacy of E7820 in Japanese patients with solid tumors. After confirming the tolerability6) of E7820 and determining its recommended dosage and administration in the phase I study, the National Cancer Center and Eisai will consider the implementation of a phase II study to assess the drug efficacy in the specified cancer types and solid tumors with the biomarkers, as well as studies for filing an application to obtain regulatory approval. At the same time, both parties will aim to further develop the system built in this research to establish a drug discovery and development system that accelerates the development of new anticancer drugs.
Background
The PDX is a type of cancer model created by directly implanting tumor tissue of cancer patients into immunodeficient mice and has been utilized in non-clinical studies and research. PDX models retain tumor heterogeneity and many of the gene mutations in the donor cancer patient and have high predictive accuracy for clinical outcomes compared to the conventional approach using cell lines and cell line-transplanted mouse models. PDX models are also reported to be highly similar to clinical data in responsiveness.
The National Cancer Center Research Institute established J-PDX library from Japanese cancer patients with relevant clinical information in 2020 and possesses 651 PDX models across different cancer types (as of July 3, 2024), which are increasingly utilized in drug discovery and development.
With the area of oncology as one of its strategic focus, Eisai strives to contribute to the cure of cancer by creating innovative new drugs with novel targets and mechanisms of action in the Deep Human Biology Learning drug discovery and development system, while making use of the experience and knowledge from globally approved in-house discovered compounds, microtubule dynamics inhibitor eribulin mesylate (product name: Halaven®) and multiple receptor tyrosine kinase inhibitor lenvatinib mesylate (product name: Lenvima®).
E7820 is an anticancer sulfonamide that acts as a molecular glue to bind ubiquin ligase DCAF15 to splicing factor RBM39, resulting in selective degradation of RBM39. This action is expected to induce antirumor efficacy by dysregulation of RNA splicing7). Clinical studies of E7820 have been conducted in foreign sites.
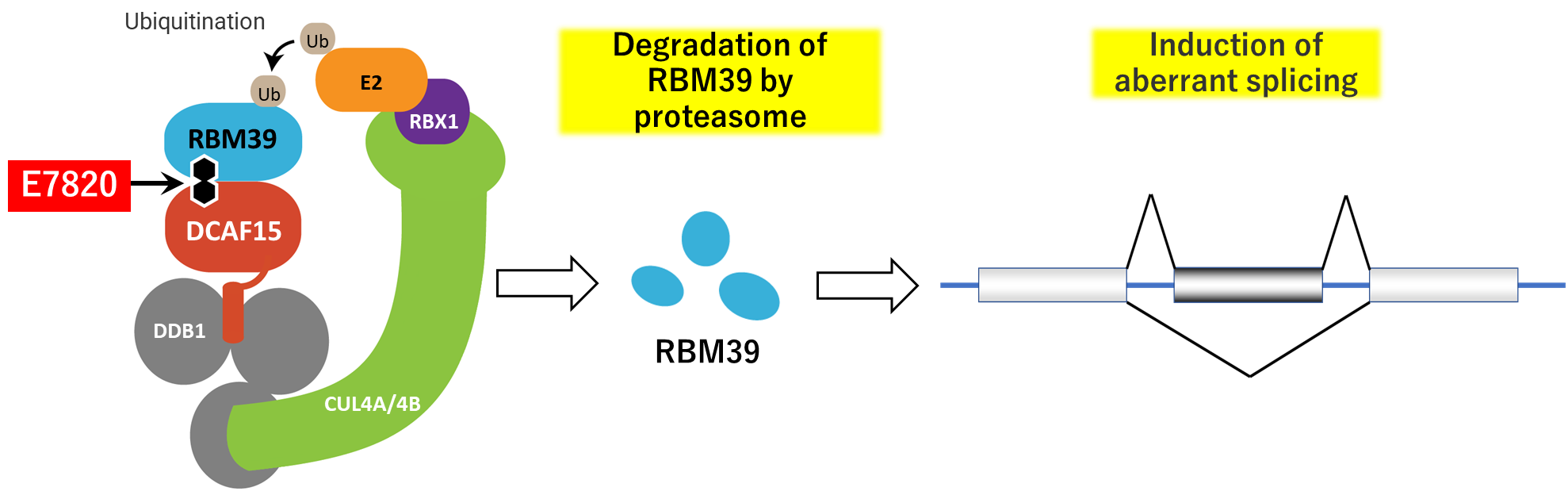
Fig. 1: E7820 bridges the binding between splicing factor RBM39 and the ubiquitin ligase DCAF15, leading to ubiquination of RBM39 and subsequrnt protasomal degradation. This induces splicing impairment, thereby exerting antitumor effects.
In the “Basic Research on the Drug Discovery and Development to Accelerate Development of Anticancer Drugs in Treatment of Patients with Rare Cancers and Refractory Cancers,” the National Cancer Center and Eisai carried out a tumor-agnostic non-clinical study of Eisai’s new drug candidate using J-PDX models and will now conduct an investigator-initiated clinical study in rare cancers and refractory cancers to confirm its clinical usefulness while aiming to apply for regulatory approval (Fig. 2). Moreover, both parties are considering expanding into new drug discovery research by using PDX models established from tumor tissue before and after treatment to conduct comparative analyses of drug responsiveness and cancer genome data to search for a new target in drug discovery and uncover the mechanisms of drug resistance. Through these efforts, both parties aim to establish a drug discovery and development system that accelerates the development of new anticancer drugs.
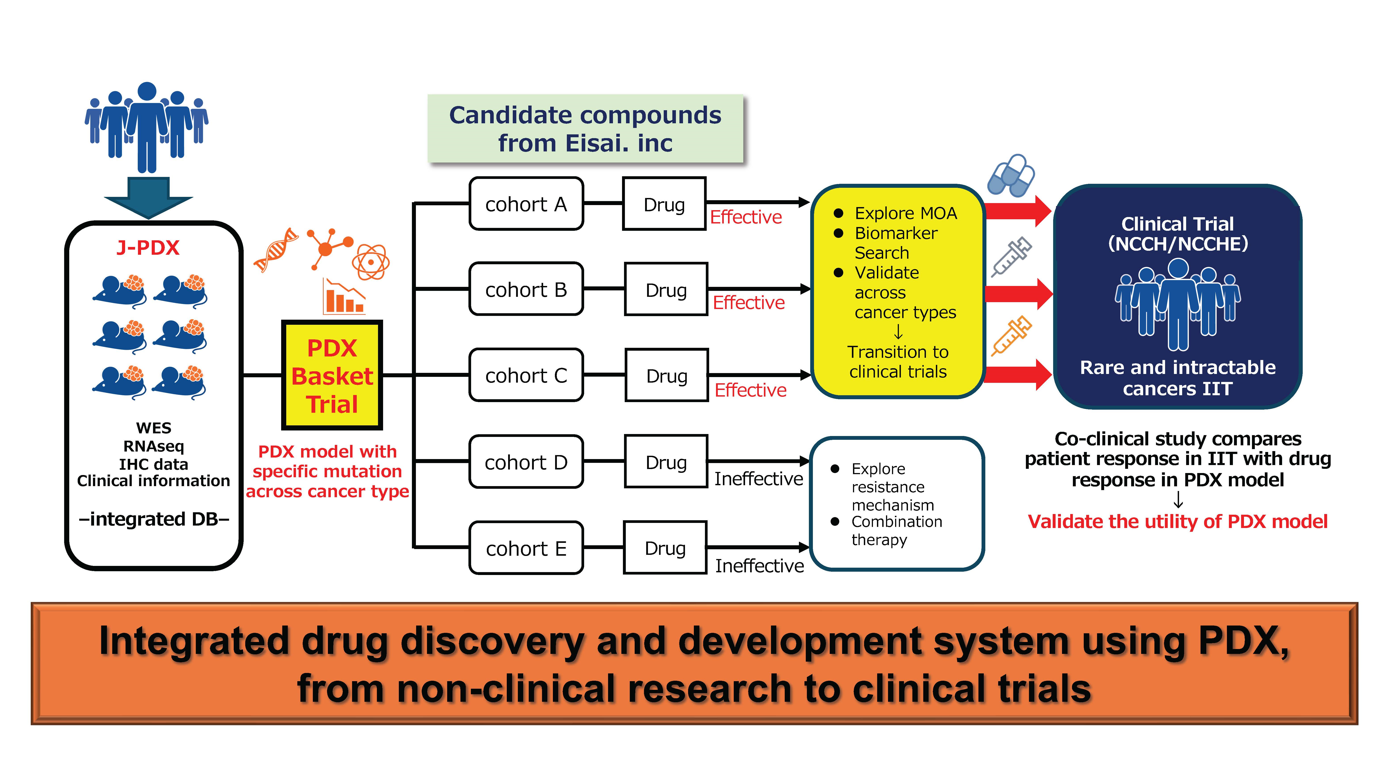
Fig. 2: Overview of the joint R&D project between the National Cancer Center and Eisai
Note May 14, 2021 Press ReleaseNATIONAL CANCER CENTER AND EISAI COMMENCE JOINT RESEARCH AND DEVELOPMENT PROJECT “BASIC RESEARCH ON THE DRUG DISCOVERY AND DEVELOPMENT TO ACCELERATE DEVELOPMENT OF ANTICANCER DRUGS IN TREATMENT OF PATIENTS WITH RARE CANCERS AND REFRACTORY CANCERS”, USING PDX WITH HIGH PREDICTABILITY OF CLINICAL OUTCOMES, AND CANCER GENOME DATA
https://www.ncc.go.jp/jp/information/pr_release/2021/0514/index.html
Study results
PDX models for evaluating the drug efficacy of E7820
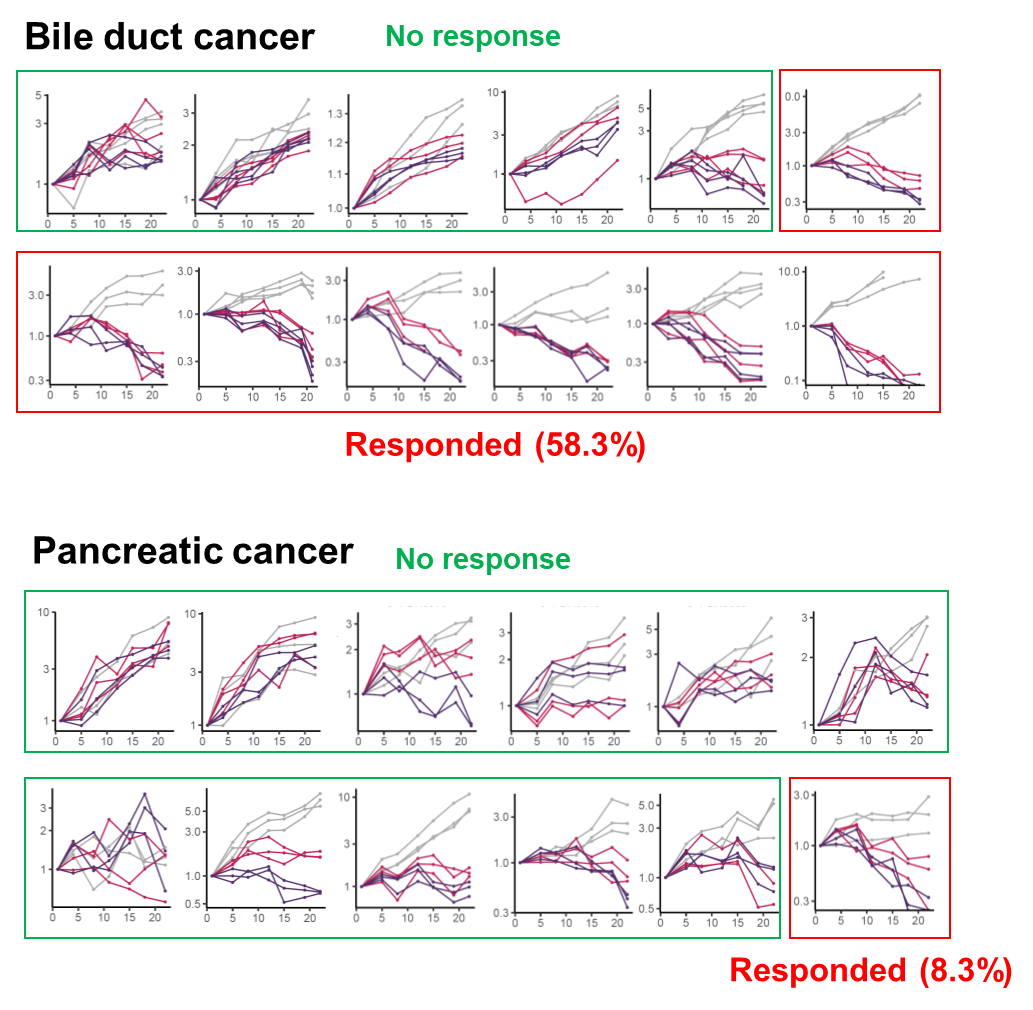
E7820 was orally administered to 42 PDX models of various types of tumors, including tumors of the bile duct (12), pancreas (12), stomach (9), and uterus (9). Significant tumor shrinkage (ΔT/C < -30%) was observed among the tumors (Fig. 3a). The overall response rates were 38.1% (16 PDX) and 54.8% (23 PDX) for 100 and 200 mg/kg of the drug administered, respectively. The highest response rate for 100 mg/kg was observed in bile duct cancer (07月12日, 58.3%), followed by uterine cancer (05月09日, 55.6%), and gastric cancer (03月09日, 33.3%). The lowest response rate was 8.3% (01月12日), in pancreatic cancer. Pancreatic cancer was significantly more resistant to treatment than other tumor subtypes (HR = 11.0, 95% confidence interval (CI) = 1.26–96.2, p = 1.5×10-2, Fisher's exact test) (Fig. 3b).
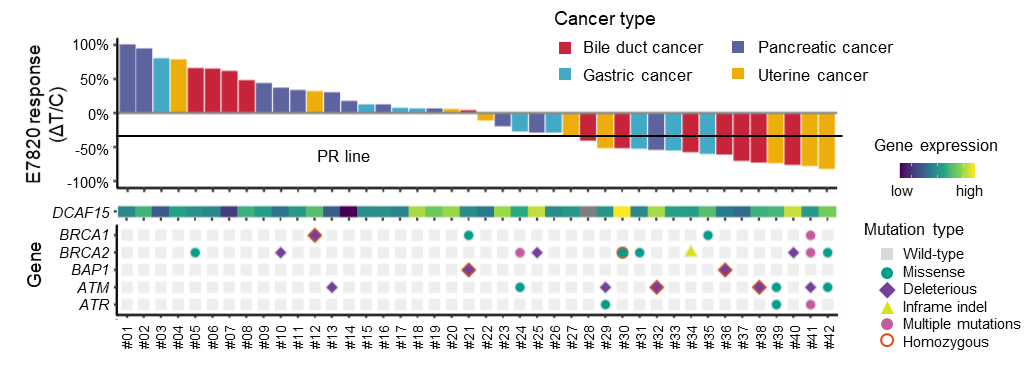
a
b
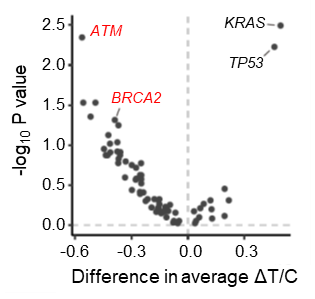
c
Fig. 3. The PDX drug evaluation screen and whole exon sequencing identified cancer types and predictive biomarkers that are associated with E7820 efficacy.
Homologous recombination deficiency (HRD) is highly observed in E7820-sensitive PDXs
WES and Whole transcriptome analysis8) were conducted to identify the molecular markers related to E7820 sensitivity. Wilcoxon's rank sum test comparing the E7820 sensitivity of PDXs with and without mutations revealed that mutations in genes related to homologous recombination repair (HRR), such as BRCA1, BRCA2 and ATM, were seemingly enriched in responders (Fig. 3a). ATM mutations were significantly enriched in responders (p = 4.5×10-3, FDR = 0.14) while BRCA2 mutations (p = 4.8×10-2, FDR = 0.51) was mildly enriched in the responder group (Fig. 3c). In contrast, TP53 was significantly mutated in non-responders (p = 5.9×10-3, FDR = 0.14), although this enrichment may be related to its high positivity in pancreatic cancer.
Cell growth inhibition becomes apparent under long-term treatment with E7820 in HRD- positive cell
A recent report indicated that E7820 affects the RNA splicing of some HRR genes but did not demonstrate that HRD can serve as a biomarker for predicting E7820 response. Since our initial screening in the PDX models showed marked shrinkage of the tumors with HRD, we hypothesized that growth suppression may become dominant after long-term exposure to the compound. Drug efficacy was thus evaluated in short-term (3 days) or long-term (12 days) cultures of cancer cell lines, including a colorectal cancer cell line with (DLD1- BRCA2-KO) or without BRCA2 knockout (DLD1-P). As shown in Fig. 4a, DLD1- BRCA2-KO cells, but not DLD1-P cells, were highly sensitive to E7820 and poly-ADP ribose polymerase inhibitor (PARPi) olaparib on day 12. To further examine whether other genes involved in the DNA damage response affect sensitivity to E7820, ATM, ATR or BAP1 genes were knocked out in DLD1 cells, which were then subjected to drug exposure. As a result, deletion of ATM, ATR or BAP1 sensitized DLD1 cells to E7820 (Fig. 4b).
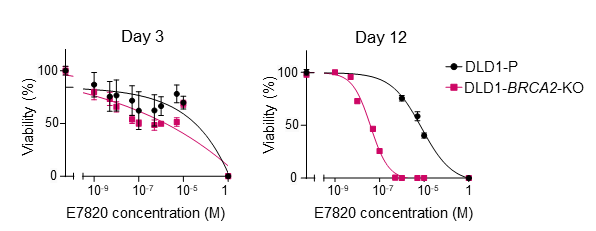
a
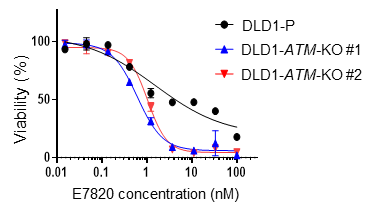
b

c
Fig. 4. (a)(b) Cell growth inhibition becomes apparent under long-term treatment with E7820 in HRD- positive cell. (c) BRCA2 dysfunction increases E7820-induced DNA double-strand breaks
BRCA2 dysfunction increases E7820-induced DNA double-strand breaks
Since DLD1- BRCA2-KO showed sensitivity to E7820, similar to olaparib, exposure to E7820 may induce DNA damage. The presence of γH2AX, a marker of DNA double-strand breaks, was evaluated in cells cultured with E7820 or olaparib, a PARP inhibitor9), for 72 h. The treatment with E7820 (1 µM) degraded RBM39 and induced the appearance of more γH2AX than treatment with olaparib (1 µM) (Fig. 4c).
Transcriptomic changes associated with E7820
The RNA-seq dataset was further evaluated to clarify how E7820 induces DNA double-strand breaks. We then isolated genes commonly upregulated or downregulated by E7820 treatment among different six cell lines. There were 1,655 upregulated genes (fold change >1.1) and 2,787 downregulated genes (fold change <0.9) observed in more than three cell lines, as annotated via ingenuity pathway analysis (IPA). Among the identified pathways shared among the downregulated genes, ““Nucleotide excision repair (NER) Enhanced Pathway,” “Hereditary Breast Cancer Signalling,” and “Role of BRCA1 in DNA Damage Response” (-log(p value) = 9.27, 8.51 and 7.38, respectively) (Fig. 5a). These pathways included HRR-related genes, such as PALB2, BRIP1, BRCA1, RAD50, MRE11, ATR and FANC family genes. Furthermore, downregulated genes also included essential genes for NER, such as XPC and ERCC2. Next, we investigated the alternative splicing events induced by E7820. In both DLD1-P and DLD1- BRCA2-KO, intron retention was the most increased splicing anomaly induced by E7820, while the most decreased splicing anomaly was single-exon skipping (Fig. 5b). Among 41 genes related to DNA repair, intron retention was induced in 25 genes (61%) in DLD1- BRCA2-KO cells (Fig. 5c).
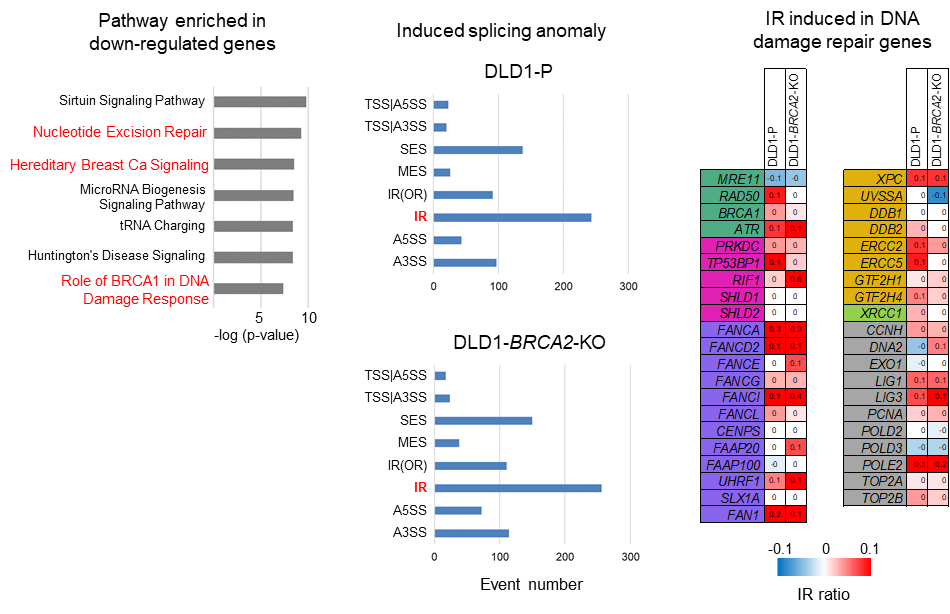
Fig. 5. Analysis of transcriptome changes associated with E7820 treatment showed decreased expression of genes involved in signaling pathways related to DNA damage repair and the induction of intron retention in those genes.
Any missplicing of mRNAs severely affects the translation of the corresponding mature proteins. Therefore, we measured the protein levels of FANCD2 and FANCA in DLD1- BRCA2-KO cells (Fig. 5d). Incubation with E7820 markedly reduced the levels of FANCD2 and FANCA proteins in a time-dependent manner. Importantly, a decrease in FANCD2/FANCA protein levels coincided with the accumulation of γH2AX and cleavage of caspase-3 (a hallmark of cell apoptosis).
Use of PDX (patient-derived xenograft) models in drug discovery
One of the challenges in anticancer drug development thus far is the low predictive ability of cell line-based experimental models. On the other hand, the PDX is an animal model in which tumor tissue of a cancer patient is implanted into an immunodeficient mouse to reproduce a tumor and is reported to have a high recall rate as it can retain the characteristic of tumor tissue. The use of PDX models in drug discovery is rapidly growing. At the National Cancer Center, the effect of anti-HER2 therapy predicted using PDX models from a uterine carcinosarcoma patient was confirmed to be consistent with clinical response to the drug.Note
The large-scale PDX library “J-PDX” derived from Japanese cancer patients built by the National Cancer Center has the following characteristics: 1) PDX models were established in a tumor-agnostic manner with the focus on rare cancers (e.g., osteosarcoma and rhabdomyosarcoma) and cancers that are common in Asia in addition to major cancers; 2) PDX models were established not just from surgical specimens but also from drug-resistant specimens; and 3) PDX models come with detailed clinical information including treatment history. Use of J-PDX in the development of new anticancer drugs would help accelerate the development process.
NoteApril 10, 2023 Press Release
Efficacy of Anti-HER2 Therapy with Trastuzumab Deluxecan in Uterine Carcinosarcoma Consistent with PDX-model predictions of efficacy, paving the way for the development of treatments for rare cancers.
https://www.ncc.go.jp/en/information/press_release/20230410/index.html
Publication
Journal: npj Precision Oncology
Title: A molecular glue RBM39-degrader induces synthetic lethality in cancer cells with homologous recombination repair deficiency
Authors: Shinji Kohsaka, Shigehiro Yagishita, Yukina Shirai, Yusuke Matsuno, Toshihide Ueno, Shinya Kojima, Hiroshi Ikeuchi, Masachika Ikegami, Rina Kitada, Ken-ichi Yoshioka, Kohta Toshimitsu, Kimiyo Tabata, Akira Yokoi, Toshihiko Doi, Noboru Yamamoto, Takashi Owa, Akinobu Hamada, Hiroyuki Mano
DOI:10.1038/s41698-024-00610-0
Publication Date: May 24, 2024
URL: https://www.nature.com/articles/s41698-024-00610-0(link to external site)
Domestic investigator-initiated phase I clinical study to evaluate the safety and efficacy of E7820 in Japanese patients with solid tumors
The dose finding part of this study will examine the tolerability, pharmacokinetics, etc. of E7820 in Japanese patients with solid tumors unresponsive or intolerant to standard therapies. The subsequent dose expansion part will evaluate the efficacy and safety of E7820 in an exploratory manner in patients with bile duct cancer, uterine cancer, or solid tumor with HRR gene mutations using the dosage and administration confirmed tolerable in the dose-finding part. The investigational product will be provided by Eisai. In overseas, clinical studies have been conducted using E7820, and the maximum tolerated dose (MTD) in once-daily oral consecutive day administration has been set at 100 mg.
Title of the study
A domestic investigator-initiated phase I clinical study to evaluate the safety and efficacy of E7820
in Japanese patients with solid tumors
(Protocol No.: NCCH2303; abbreviated study title: CIRCUS)
drug used
E7820 (oral drug: splicing inhibitor sulfonamide)
Physical condition of patients eligible to participate (inclusion criteria)
- The patient can confirm consent in written form.
- The patient has a solid tumor diagnosed as unresectable advanced or recurrent cancer.
- No standard therapies are available for the cancer, or the cancer has been determined to be refractory or intolerant to standard therapies.
- The patient is 18 years old or older at the time of enrollment.
- Functions of each organ in the patient are maintained within the specified range.
Note: Please note that the above is an outline of the inclusion criteria and that some patients who satisfy all of the above may still not be eligible to participate in this study.
Principal Investigator
Noboru Yamamoto
Chief, Department of Experimental Therapeutics, National Cancer Center Hospital
Sites
National Cancer Center Hospital
National Cancer Center Hospital East
Japan Registry of Clinical Trials
jRCT No.: jRCT2031240210
For details on this clinical study, please visit the Japan Registry of Clinical Trials website.
https://jrct.niph.go.jp/latest-detail/jRCT2031240210(link to external site)
Research funding
Cyclic Innovation for Clinical Empowerment(CiCLE) by Japan Agency for Medical Research and Development(AMED)(JP20pc0101051)
Prospects
By using the National Cancer Center’s J-PDX library in company-led drug discovery and development, it has become possible to evaluate the efficacy of a new drug candidate, identify predictive biomarkers associated with the drug efficacy, and launch a clinical study in a seamless manner. After confirming the safety of E7820 including its tolerability and determining the recommended dose in this study, the National Cancer Center and Eisai will consider the implementation of phase II study to evaluate the efficacy of the drug in the specified cancer types and solid tumors with the biomarkers, as well as studies for filing an application to obtain regulatory approval.
At the same time, both parties will aim to further develop the system built in this research to establish a drug discovery and development system that accelerates the development of new anticancer drugs.
Glossary
1) PDX (patient-derived xenograft): mouse model
A type of cancer model created by directly transplanting tumor tissue of cancer patients into immunodeficient mice.
2) Biomarkers
Molecules such as proteins and genes that can be an indicator for the presence of a disease or treatment selection.
3) Whole-exome sequencing
A technique to selectively enrich and efficiently analyze exons that encode proteins in the human genome.
4) Homologous recombination repair (HRR)
One of the main mechanisms for repairing DNA double-strand breaks. Uses a sister chromatid as a template for repair.
5) Investigator-initiated clinical studies
For a new drug to be approved and covered by health insurance, it requires clinical development (clinical studies). In the case where no clinical studies are conducted by a company for a promising drug, physicians may carry out clinical studies for the drug with the cooperation of a pharmaceutical company, which is called investigator-initiated clinical studies.
6) Tolerability
A term representing the degree to which adverse effects of an investigational product can be tolerated by humans. Specifically, factors such as the incidence and severity of adverse effects considered intolerable in humans are taken into account.
7) RNA splicing
An essential mechanism for gene expression that removes introns from precursor mRNA to produce mature mRNA.
8) Whole transcriptome analysis
A technique to analyze the sequences and the amount of gene expression of the entire gene transcripts in a cell (transcriptome).
9) PARP inhibitors
Drugs that inhibit the enzyme poly ADP-ribose polymerase (PARP), which plays a vital role in cells such as DNA repair, to induce synthetic lethality in tumor cells with mutations in homologous recombination repair genes.
Contact
- Non-clinical research Inquiries
Shinji Kohsaka, Chief, Division of Cellular Signaling, National Cancer Center Research Institute
E-mail: skohsakancc.go.jp - Investigator-initiated clinical trial Inquires
International Trial Management Section, Research Management Division, Clinical Research Support Office, National Cancer Center Hospital
E-mail: ncch2303_officeml.res.ncc.go.jp - Media Inquiries
Office of Public Relations, Strategic Planning Bureau, National Cancer Center
E-mail:ncc-adminncc.go.jp
Eisai Co., Ltd. Public Relations Department
TEL : +81-(0)3-3817-5120
